| (19) |
 |
|
(11) |
EP 0 515 698 B1 |
| (12) |
EUROPEAN PATENT SPECIFICATION |
| (45) |
Mention of the grant of the patent: |
|
29.07.1998 Bulletin 1998/31 |
| (22) |
Date of filing: 06.12.1991 |
|
| (86) |
International application number: |
|
PCT/JP9101/696 |
| (87) |
International publication number: |
|
WO 9210/579 (25.06.1992 Gazette 1992/14) |
|
| (54) |
Process for producing D-alpha-amino acids
Verfahren zur Herstellung von D-Alpha-Aminosäuren
Procédé de production de D-alpha aminoacides
|
| (84) |
Designated Contracting States: |
|
BE DE ES FR GB IT NL |
| (30) |
Priority: |
07.12.1990 JP 400848/90
27.12.1990 JP 407922/90
11.04.1991 JP 78840/91
|
| (43) |
Date of publication of application: |
|
02.12.1992 Bulletin 1992/49 |
| (73) |
Proprietor: KANEGAFUCHI KAGAKU KOGYO KABUSHIKI KAISHA |
|
Kita-ku
Osaka-shi
Osaka-fu 530 (JP) |
|
| (72) |
Inventors: |
|
- NANBA, Hironori
Takasago-shi,
Hyogo 676 (JP)
- YAMADA, Yukio
Kakogawa-shi,
Hyogo 675 (JP)
- TAKANO, Masayuki,
106, Leopalace
Futami-cho,
Akashi-shiHyogo 674 (JP)
- IKENAKA, Yasuhiro
Akasi-shi,
Hyogo 673 (JP)
- TAKAHASHI, Satomi
Kobe-shi,
Hyogo 655 (JP)
- YAJIMA, Kazuyoshi
Suma-ku,
Kobe-shi,
Hyogo 654-01 (JP)
|
| (74) |
Representative: Kolb, Helga, Dr. Dipl.-Chem. et al |
|
Hoffmann Eitle,
Patent- und Rechtsanwälte,
Postfach 81 04 20
81904 München
81904 München (DE) |
| (56) |
References cited: :
EP-A- 0 136 359
GB-A- 1 534 426
JP-A-63 024 894
JP-B-57 018 793
|
EP-A- 0 261 836
GB-A- 2 022 581
JP-A-63 185 382
|
|
| |
|
|
- PATENT ABSTRACTS OF JAPAN vol. 12, no. 230 (C-508)(3077) 29 June 1988 & JP-A-63 024
894 (AJINOMOTO CO INC) 2 February 1988
- BIOTECHNOLOGY AND BIOENGINEERING. vol. 23, no. 10 , 1981 , NEW YORK US pages 2173
- 2183 R. OLIVIERI ET AL 'Microbial transformation of racemic hydantoins to d-amino
acids'
|
|
| |
|
| Note: Within nine months from the publication of the mention of the grant of the European
patent, any person may give notice to the European Patent Office of opposition to
the European patent
granted. Notice of opposition shall be filed in a written reasoned statement. It shall
not be deemed to
have been filed until the opposition fee has been paid. (Art. 99(1) European Patent
Convention).
|
Technical field
[0001] The present invention relates to a process for the production of D-α-amino acids,
and more particularly, to a process for the production of D-α-amino acids using a
novel transformant having a gene which is related to an enzyme capable of converting
D-N-carbamoyl-α-amino acids into the corresponding D-α-amino acids.
Background art
[0002] Optically-active D-α-amino acids are important compounds as intermediates of drugs,
and particularly, D-phenylglycine, D-parahydroxyphenyl-glycine and other intermediates
for the production of semisynthesized penicillin or cephalosporin antibiotics are
industrially useful compounds. As a process for the production of such D-α-amino acids,
there is a well-known process in which carbamoyl groups of the corresponding D-N-carbamoyl-α-amino
acids are removed to give the desired D-α-amino acids. The removal of carbamoyl groups
in this process is achieved by a chemical process (e.g., the specification of Japanese
Patent Publication No. 58-4707) or by a process utilizing the enzymatic reaction of
microorganisms (e.g., the specifications of Japanese Patent Publication Nos. 57-18793,
63-20520, and 1-48758).
Problems to be solved by the invention
[0003] In a chemical process employed for the removal of carbamoyl groups as described above,
a great amount of mineral acid such as sulfuric acid is used, and therefore, there
will occur serious environmental problems regarding to the disposal thereof and the
like. On the other hand, a process utilizing the enzymatic reaction of microorganisms
has several drawbacks that microorganisms hitherto known as a source of enzyme supply
cannot produce a sufficient amount of enzymes and that expensive hydantoin or N-carbamoylamino
acid compounds are required for the production of enzymes.
Means for solving the problems
[0004] For the purpose of solving such problems, the objects of the present invention are
to prepare microorganisms having high productivity of enzymes, as well as to produce
D-α-amino acids with high efficiency by use of a source of enzyme supply thus obtained.
[0005] A similar technique is disclosed in Japanese Patent Laid-open Publication No. 63-24894.
However, this technique relates to the production of L-α-amino acids, and there is
no experimental example describing the production of D-α-amino acids.
[0006] The present invention provides a process for the production of D-α-amino acids, by
a method in that D-N-carbamoyl-α-amino acids are converted into the corresponding
D-α-amino acids in an aqueous medium with the aid of an action of an enzyme capable
of converting D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into
the corresponding D-α-amino acids, characterized in that said enzyme is produced by
a transformant which is obtainable by transformation of host bacterial cells selected
from the microorganisms belonging to the genera
Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium, and
Brevibacterium, with a recombinant DNA comprising a vector DNA and a DNA fragment containing a gene
encoding said enzyme after which the D-α-amino acids produced are collected.
[0007] By the way, no example has hitherto been known that a recombinant DNA comprising
a vector and a gene which is related to an enzyme capable of converting D-N-carbamoyl-α-amino
acids by removal of their carbamoyl groups into the corresponding D-α-amino acids
is incorporated into microorganisms to achieve the expression of the gene. Such a
technique was not succeeded until the present invention has been completed.
[0008] The enzymes capable of converting D-N-carbamoyl-α-amino acids by removal of their
carbamoyl groups into the corresponding D-α-amino acids are, in fact, not limited
to those which specifically act on the D-isomers or those which act on either D- or
L-isomers. In particular, enzymes having a strict stereoselectivity to D-N-carbamoyl-α-amino
acids may be referred to as D-N-carbamoyl-α-amino acid amidohydrolases
[0009] Examples of the DNA fragment containing a gene which can be used in the present invention
are those which are derived from eucaryotes, prokaryotes, viruses, bacteriophages
or plasmids, and which contain a gene related to a particular D-N-carbamoyl-α-amino
acid amidohydrolase. As the gene derived from prokaryotes, preferred are those which
are derived from bacteria belonging to the genus, for example,
Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium, Serratia, Micrococcus, Arthrobacter, Alkaligenes, Achromobacter, Moraxella, or
Paracoccus, and which are related to D-N-carbamoyl-α-amino-acid amidohydrolases. Specific examples
of such strains are as follows:
Aerobacter cloacae IAM 1221,
Bacillus macroides ATCC 12905,
Bacillus alvei IFO 3343,
Brevibacterium ammoniagenes IFO 12071,
Flavobacterium flavescens IFO 3086,
Sarcina lutea IFO 1099,
Serratia marcescens IFO 305
4,
Micrococcus luteus IFO 12708,
Aeromonas hydrophilia IFO 3820,
Agrobacterium species KNK 712 (FERM BP-1900),
Pseudomonas-sp. KNK 003A (FERM BP-3181),
Pseudomonas sp. KNK 505 (FERM BP-3182),
Agrobacterium 1302 NRRL B11291,
Alkaligenes aquamarinus AJ 11199,
Achromobacter liquefaciens AJ 11198,
Moraxella nonliquefaciens AJ 11221,
Paracoccus denitrificans AJ 11222,
Arthrobacter fragilus AJ 11223, and the like.
[0010] As the typical examples of the above bacteria, the microbiological characteristics
of
Agrobacterium species KNK 712 (FERM BP-1900),
Pseudomonas sp. KNK 003A (FERM BP-3181), and
Pseudomonas sp. KNK 505 (FERM BP-3182) are described below.
Agrobacterium species KNK 712 (FERM BP-1900)
[0011]
(a) Morphology
(1) Cell size and shape: 0.5-1.0 x 2.0-4.0 µm, rods
(2) Cellular polymorphism: none
(3) Motility and flagellar arrangement: active, subpolar
(4) Sporulation: none
(5) Gram-staining: negative
(b) Cultural characteristics on various media
(1) Meat extract agar plate culture:
satisfactory growth, round, ridgy, smooth margin, smooth wet surface, white to
cream-colored, glossy, opaque, liquid form
(2) Meat extract agar slant culture:
excellent growth, even growth on the inoculation line,- thick ridge, smooth wet
surface, smooth margin, white to cream-colored, glossy, opaque, no change in the medium
(3) Meat extract gelatin stab culture:
feeble growth, growth along the stab line, growth around the stab line, no gelatin
liquefaction, white to cream-colored, no change in transparency, no change in the
medium
(4) Meat extract liquid culture:
medium growth, uneven turbidity, flock formation, no surface growth
(5) Litmus milk: weak acid
(c) Physiological Characteristics
(1) Nitrate reduction: +
(2) Denitrification: +
(3) MR test: +
(4) Vp test: -
(5) Indole formation: -
(6) Hydrogen sulfide formation: -
(7) Starch hydrolysis: -
(8) Citrate utilization: - (Simmons medium)
(9) Inorganic nitrogen source: + (nitrates, ammonium salts)
(10) Urease: -
(11) Oxidase: +
(12) Catalase: +
(13) Growth range: 20-37°C, pH 6.5-8.5
(14) Attitude toward oxygen: aerobic
(15) O-F test: O type
(16) Formation of acid and gas from saccharide: Acid formation, -; Gas formation,
- (D-glucose)
(17) Malonate utilization: -
(18) Deaminase reaction of phenylalanine: -
(19) Decarboxylase reaction: - (lysine)
(20) Arginine dihydrolase reaction: -
(21) Casein degradation: -
(22) DNA degradation: -
(23) Auxotrophy: none
(24) Utilization of carbon compounds:
D-Glucose: +
L-Arabinose: +
Saccharose: +
D-Fructose: +
Malonate: -
Cellobiose: +
Ethanol: -
D-Xylose: +
D-Tartrate: -
Sorbitol: +
Citrate: -
Lactose: +
D-Mannitol: +
Meso-inositol: +
Raffinose: +
L-Rhamnose: +
Maltose: +
α-Methyl-D-glucoside: +
D-Mannose: +
Salicin: -
N-Acetylglucosamine: +
Gluconate: -
Caprate: -
Adipate: -
Phenylacetate: -
Methanol: -
(25) Egg york reaction:-
(26) β-Galactosidase: +
(27) Esculin hydrolysis: +
(28) Cytochrome oxidase: +
(29) Tween® degradation: - (Tween® 80)
(30) 3-Ketolactose formation: -
Pseudomonas sp. KNK 003A (FERM BP-3181)
[0012]
(a) Morphology:
(1) Cell size and shape: 0.5-0.7 × 1.2-2.5 µm, rods
(2) Cellular polymorphism: none
(3) Motility: active
(4) Sporulation: none
(5) Gram-staining: negative
(6) Colony shape: round, regular, entire, flat, smooth, shiny, semi-translucent, buff
(b) Physiological Characteristics
(1) Lysis by 3% KOH: +
(2) Aminopeptidase: +
(3) Oxidase: +
(4) Catalase: +
(5) Cell growth
Anaerobic condition: -
37/40°C: +/+
pH 5.6: -
Mac-Conkey agar: -
SS agar: -
Cetrimid agar: -
(6) Acid formation (O-F test)
Glucose aerobic condition: -
Glucose anaerobic condition: -
(7) Gas formation from glucose: -
(8) Acid formation (ASS)
Glucose: +
Fructose: -
Xylose: +
(9) ONPG: -
(10) ADH: -
(11) ODC: -
(12) VP: -
(13) Indole formation: -
(14) Nitrate reduction: -
(15) Denitrification: -
(16) Phenylalanine deaminase: -
(17) Levan formation from sucrose: -
(18) Lecithinase: -
(19) Urease: -
(20) Hydrolysis
Starch: -
Geratin: -
Casein: -
DNA: -
Tween® 80: -
Esculin: -
(21) Tyrosine degradation: -
(22) Utilization of various compounds
Acetate: weak
Adipate: -
Caprate: -
Citrate: -
Citraconate: -
Glycolate: -
Lactate: +
Levulinate: -
Malate: -
Malonate: -
Mesaconate: -
Phenylacetate: -
Suberate: -
m-Tartrate: -
D-Tartrate: -
L-Arabinose: +
Fructose: +
Glucose: +
Mannose: +
Maltose: -
Xylose: +
Saccharose: -
Trehalose: -
Ribose: -
Saccharate: -
Hydroxybutylate: -
Benzoate: -
Mannitol: +
Gluconate: +
2-Ketogluconate: +
N-Acethylglucosamine: -
L-Serine: -
L-Histidine: -
L-Valine: -
(23) Main respiratory quinone type: ubiquinone 10
Pseudomonas sp. KNK 505 (FERM BP-3182)
[0013]
(a) Morphology:
(1) Cell size and shape: 0.5-0.7 × 1.2-2.5 µm, rods
(2) Cellular polymorphism: none
(3) Motility: active
(4) Sporulation: none
(5) Gram-staining: negative
(6) Colony shape: round, regular, entire, flat, smooth, shiny, semi-translucent, buff
(b) Physiological Characteristics
(1) Lysis by 3% KOH: +
(2) Aminopeptidase: +
(3) Oxidase: +
(4) Catalase: +
(5) Cell growth
Anaerobic condition: -
37/40°C: +/+
pH 5.6: -
Mac-Conkey agar: -
SS agar: -
Cetrimid agar: -
(6) Acid formation (O-F test)
Glucose aerobic condition: -
Glucose anaerobic condition: -
(7) Gas formation from glucose: -
(8) Acid formation (ASS)
Glucose: +
Fructose: -
Xylose: +
(9) ONPG: -
(10) ADH: -
(11) ODC: -
(12) VP: -
(13) Indole formation: -
(14) Nitrate reduction: -
(15) Denitrification: -
(16) Phenylalanine deaminase: -
(17) Levan formation from sucrose: -
(18) Lecithinase: -
(19) Urease: -
(20) Hydrolysis
Starch: -
Geratin: -
Casein: -
DNA: -
Tween® 80: -
Esculin: -
(21) Tyrosine degradation: -
(22) Utilization of various compounds
Acetate: weak
Adipate: -
Caprate: -
Citrate: -
Citraconate: -
Glycolate: -
Lactate: +
Levulinate: -
Malate: -
Malonate: -
Mesaconate: -
Phenylacetate: -
Suberate: -
m-Tartrate: -
D-Tartrate: -
L-Arabinose: +
Fructose: +
Glucose: +
Mannose: +
Maltose: -
Xylose: +
Saccharose: -
Trehalose: -
Ribose: -
Saccharate: -
Hydroxybutylate: -
Benzoate: -
Mannitol: +
Gluconate: +
2-Ketogluconate: +
N-Acethylglucosamine: -
L-Serine: -
L-Histidine: -
L-Valine: -
(23) Main respiratory quinone type: ubiquinone 10
[0014] To obtain a gene from these strains, which is related to an enzyme capable of converting
D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into the corresponding
D-α-amino acids, usually, genetic DNA is extracted from the chromosome of microorganisms
according to the conventional procedure, after which a DNA fragment containing the
desired gene is obtained and subjected to an analysis for its base sequence. Moreover,
microorganisms which produce an enzyme capable of converting D-N-carbamoyl-α-amino
acids by removal of their carbamoyl groups into the corresponding D-α-amino acids,
as well as transformed microorganisms into which this enzyme gene has been incorporated,
are cultivated, and the produced enzyme is purified, after which the molecular weight
of its protein is determined and the amino acid sequence in the vicinity of its amino
terminus is determined by a gas-phase protein sequencer or the like. Then, this DNA
base sequence is compared with the amino terminal sequence of the protein, so that
the initiation site for genetic translation into a protein, of the base sequence portion
encoding an enzyme protein which is related to removal of carbamoyl groups is determined,
and taking into consideration the relation to the molecular weight of the protein,
it is confirmed that the enzyme protein is encoded in the gene portion extending from
this site to the termination codon, thereby verifying the desired gene (Molecular
Cloning, A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Chaps.
4 and 13). According to these procedures, DNA fragments of SEQ ID Nos. 1 and 2 in
the accompanying Sequence Listing were obtained from the strains
Agrobacterium species KNK 712 and
Pseudomonas sp. KNK 003A. The thus-obtained gene encoding the above enzyme-and/or DNA fragment
containing this gene are equivalent to DNA fragments having another base sequence
which encodes an amino acid sequence corresponding to the above gene and/or DNA fragment
because one amino acid usually corresponds to a plurality of base codons, and this
fact is obvious.
[0015] As the vector used in the present invention, plasmids, phages, or derivatives thereof
can be used, which are derived from microorganisms and can be autonomously grown in
cells of a bacteria belonging to the genus
Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Croynebacterium, or
Brevibacterium. For example, host-vector systems described in "Guidelines on Recombinant DNA Experiments"
(Ed., the Life Science Section of the Research Development Office-in the Science and
Technology Agency: revised on September 16, 1987), page 55, can be used. Moreover,
vectors, which have been modified to have a strong structural promoter for the purpose
of increasing the amount of enzyme to be produced, can also be used.
[0016] The preparation of a recombinant comprising a vector DNA and a gene-containing DNA
fragment can be conducted by freely using the known
in vitro recombinant DNA technique. The
in vitro DNA recombination is usually conducted by cleavage and ligation (ligase reaction)
of a vector DNA and a donor DNA containing the desired gene (e.g., see the specifications
of Japanese Patent Application No. 56-211908 and United States Patent No. 4,237,224).
Many kinds of recombinant DNA, in addition to the desired recombinant DNA, are produced
by ligase reaction, and therefore, for the purpose of selectively obtaining the desired
recombinant DNA, microorganisms belonging to the genus
Escherichia,
Pseudomonas,
Flavobacterium,
Bacillus,
Serratia,
Croynebacterium, or
Brevibacterium may be directly transformed with a ligase reaction mixture, and the resulting transformants
on which the inherited character from the genetic information of the desired gene
has been conferred may be selectively separated, and the desired recombinant DNA may
be extracted and isolated from their cultured bacterial cells.
[0017] For example, the recombinant can also be obtained without direct transformation of
bacterial species belonging to any one of the above genera; that is, the desired gene
is once cloned in a host vector system using other microorganisms, such as
Escherichia coli, after which recombinant DNA with an appropriate vector is
in vitro produced, and then the above bacterial species are transformed, followed by selective
separation of transformants in the same manner as described above.
[0018] The description of the following documents can be widely applied to the production
of recombinants: the specification of United States Patent No. 4,237,224 to S.N. Cohen,
et al.; "Idenshi-sousa Jikken-hou (Experimental Method of Gene Manipulation)" [Ed.,
Yasuyuki Takagi, Kohdansha Scientific (1980)]; Method in Enzymdogy,
68, Recombinant DNA [Ed., Ray Mv, Academic Press (1979)]; the specification of Japanese
Patent Application No. 56-211908, etc.
[0019] In the case of
Escherichia coli transformed with various kinds of recombinant DNA, a method for selecting, from the
transformed strains, particular transformed strains which have the desired gene, i.e.,
the gene of a particular D-N-carbamoyl-α-amino acid amidohydrolase, and in which that
gene has been expressed, is conducted as follows: colonies of the transformed strains
are first grown on the plate containing a selection marker, such as ampicillin. Then,
various colonies of these transformed strains having the recombinant DNA are collected
and suspended in physiological saline, after which this suspension is inoculated on
the minimum liquid medium containing a particular D-N-carbamoyl-α-amino acid as a
sole nitrogen source. On this medium, only transformed strains capable of utilizing
the D-N-carbamoyl-α-amino acid, i.e., transformed strains on which D-N-carbamoyl-α-amino
acid amidohydrolase activity has been now conferred, can be grown. The culture solution
thus obtained is inoculated on the above minimum medium, and such an operation is
repeated, resulting in an enrichment of the desired transformed strains. From this
enriched culture solution, bacterial cells are separated according to the conventional
method, and the separated transformed strains are cultivated to make a bacterial reaction
with a D-N-carbamoyl-α-amino acid as a substrate, whereby the transformed strains
containing the desired gene can be obtained by confirming the production of a D-α-amino
acid.
[0020] From the transformed strains thus obtained, recombinant DNA is extracted according
to the conventional method, such as a method using alkali denaturation (see Molecular
Cloning A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Chap. 1);
the structural gene of the desired enzyme in the DNA fragments containing the cloned
desired gene is subcloned and unnecessary DNA is removed; recombinant DNA modified
to have a strong promoter is prepared in a vector; the above host bacteria are transformed
therewith; and the resulting transformed strains are used to increase the amount of
the desired enzyme to be produced.
[0021] The introduced character of the recombinant DNA can be expressed by cultivating the
transformed strains on a conventional nutrient medium. In cases where the character
from the gene DNA or vector DNA is conferred on the recombinant DNA, any drug may
be supplemented to the medium depending upon that character.
[0022] To take the transformed strains thus obtained, as a source of enzyme supply, culture
may be prepared with the use of a conventional medium, and when necessary, it is also
possible to conduct a treatment for enzyme induction, such as an addition of hydantoin
compounds, D-N-carbamoyl-α-amino acids, isopropyl-1-thio-β-D-galactosides (IPTG),
or the like, and a temperature increase.
[0023] Usually, the medium used for cultivating the transformed strains may be a conventional
medium containing carbon sources, nitrogen sources, and inorganic ions. If organic
tracenutrients, such as vitamines and amino acids, are added thereto, satisfactory
results may be obtained in many cases. As the carbon sources, carbohydrates, such
as glucose and sucrose; organic acids, such as acetic acid; alcohols, and the like,
are conveniently used. As the nitrogen sources, ammonia gas, ammonia water, ammonium
salts, and the like, are used. As the inorganic ions, phosphate ion, magnesium ion,
potassium ion, iron ion, and the like, may be used.
[0024] If culture is prepared under aerobic conditions for 1-10 days, while adjusting the
pH and temperature to an appropriate range of 4-8 and 25-45°C, respectively, it is
possible to obtain desirable results.
[0025] Examples of the embodiments acting as an enzyme produced by the transformed strains
include culture solutions of the transformed strains, bacterial cells, treated bacterial
cells, enzymes extracted from bacterial cells, immobilized bacterial cells, and the
like.
[0026] As the bacterial cells, any of the culture solution as it is after completion of
cultivation, bacterial cells separated from the culture solution, washed bacterial
cells, and the like, can be used. As the treated bacterial cells, lyophilized bacterial
cells, acetone-dried bacterial cells, bacterial cells brought into contact with toluene
or detergents, lysozyme-treated bacterial cells, bacterial cells exposed to ultrasonic
waves, mechanically ground bacterial cells, and the like, can be used, as well as
enzyme extracts having the enzyme activity to convert D-N-carbamoyl-α-amino acids
obtained from these treated bacterial cells, by removal of their carbamoyl groups
into the corresponding D-α-amino-acids; these immobilized bacterial cells; insolubilized
treated bacterial cells; enzyme proteins fixed on a support for immobilization (e.g.,
anion exchange resin), and the like. For an immobilization method, for example, it
is possible to make reference to the specification of Japanese Patent Laid-open Publication
No. 63-185382.
[0027] As the support used for immobilization, suited are phenol-formaldehyde anion exchange
resins, such as Duolite A568 or DS17186 (Rohm & Haas Co.: registered trade mark);
and various anion exchange resins containing various amines, ammonium salts, or functional
groups of the diethanolamine type, for example, polystylene resins, such as Amberlite
IRA935, IRA945, IRA901 (Rohm & Haas Co.: registered trade mark), Lewatit OC1037 (Bayer
A.G.: registered trade mark), and Diaion EX-05 (Mitsubishi Chemical Industries Ltd.:
registered trade mark). Other supports, such as DEAE-cellulose, can also be used.
[0028] Further, to obtain stronger and more stable adsorption of enzymes, usually, cross-linking
agents are used, the preferred examples thereof being glutaraldehyde. As the enzyme
to be used, not only purified enzymes, but also those with different degrees of purification,
such as partially purified enzymes, suspensions of disintegrated bacterial cells,
and cell-free extracts, can be used.
[0029] The preparation of immobilized enzymes can be conducted by using an enzyme solution
and the conventional method in which, for example, enzymes are adsorbed on a support,
followed by cross-linking treatment.
[0030] D-N-carbamoyl-α-amino acids, which are used as a substrate of the enzymatic reaction
of the present invention, can be represented by the formula: R-CH(NHCONH
2)-COOH, and as a practical embodiment, in cases where the enzyme used has strict stereoselectivity
to D-N-carbamoyl-α-amino acids, it can be used as the D-form or a mixture of the D-
and L-forms. Moreover, in cases where the stereo-selectivity is not strict because
enzymes also act on L-carbamoyl amino acids, or where enzymes are used as an enzyme
mixture also acting on the L-form thereof, it is preferred that only enzymes of the
D-form are used to produce α-amino acids of the D-form.
[0031] The substituent R can be selected in a wide range, as described in the specifications
of Japanese Patent Publication Nos. 57-18793, 63-20520, and 1-48758; in particular,
to provide industrially useful compounds, such as intermediates of drugs, it is preferred
that R is phenyl, phenyl substituted with hydroxy, alkyl, substituted alkyl, aralkyl,
or thienyl. In the case of phenyl substituted with hydroxy, the number of hydroxy
is one or more, and they may be attached at any of o-, m-, and p-positions, with the
typical example thereof being p-hydroxyphenyl. The alkyl group is such a group of
1-4 carbon atoms that the corresponding amino acid becomes D-alanine, D-valine, D-leucine,
D-isoleucine, or-the like. The substituted alkyl group is such an alkyl having 1-4
carbon atoms substituted with hydroxy, alkylthio, carboxyl, amino, phenyl, phenyl
substituted with hydroxy, amido, or the like, that the corresponding amino acid becomes
D-serine, D-threonine, D-methionine, D-cysteine, D-asparagine, D-glutamine, D-tyrosine,
D-tryptophane, D-aspartic acid, D-glutamic acid, D-histidine, D-lysine, D-arginine,
D-citrulline, or the like. The aralkyl group is such a group having 7-8 carbon atoms,
for example, benzyl or phenyl, that the corresponding amino acid becomes D-phenylalanine
or the like.
[0032] As the aqueous medium, those containing water, buffers, or organic solvents, such
as ethanol, can be used. Further, when necessary, nutrients required for the growth
of microorganisms, antioxidants, detergents, coenzymes, hydroxylamines, metals, or
the like, can also be added to the aqueous medium.
[0033] In cases where, while cultivating the bacterial cells of the above microorganisms
in a water-soluble medium, the bacterial cells are brought into contact with a particular
D-N-carbamoyl-α-amino acid, an aqueous medium containing not only a D-N-α-amino acid
but also nutrients required for the growth of microorganisms, such as carbon sources,
nitrogen sources, and inorganic ions, is used. Further, if organic tracenutrients,
such as vitamines and amino acids, are added thereto, satisfactory results can be
obtained in many cases. As the carbon sources, carbohydrates, such as glucose and
sucrose; organic acids, such as acetic acid; alcohols, and the like, are conveniently
used. As the nitrogen sources, ammonia gas, ammonia water, ammonium salts, and the
like, are used. As the inorganic ions, phosphate ion, magnesium ion, potassium ion,
iron ion, and the like, may be used.
[0034] The cultivation is conducted under aerobic conditions, while adjusting the pH and
temperature to an appropriate range of 4-8 and 25-45°C, respectively. If the cultivation
is conducted for 1-10 days, D-N-carbamoyl-α-amino acids can be converted into only
D-α-amino acids with high efficiency.
[0035] To the contrary, in cases where the culture solution of the above microorganisms
is allowed as it is, to react with cultivated bacterial cells, treated bacterial cells,
enzyme extracts, immobilized bacterial cells, insolubilized bacterial cells, or fixed
enzyme proteins, in an aqueous medium containing a particular D-N-carbamoyl-α-amino
acid dissolved or suspended, the reaction mixture may be allowed to stand for some
time or stirred, while maintaining at an appropriate temperature of 10-80°C and pH
of 4-9.5. Thus, if 5-100 hours have run their course, the corresponding D-α-amino
acid is produced in large quantities and enriched in the aqueous medium. Moreover,
the D-N-carbamoyl-α-amino acid may be added in separate portions with the progress
of reaction. The produced D-α-amino acid can be separated and purified by a conventional
separation method.
[0036] The D-α-amino acid thus obtained can be represented by the formula: R-CHNH
2COOH (R is as defined above).
Examples
[0037] The following will describe the embodiments of the present invention. The D-α-amino
acid produced was detected and determined by high performance liquid chromatography
(HPLC) or thin layer chromatography (TLC).
Example 1
[0038] Preparation of recombinant DNA comprising chromosome DNA from
Agrobacterium species KNK 712 (FERM BP-1900) and vector DNA:-
[0039] Agrobacterium species KNK 712 (FERM BP-1900) was cultivated in 2 liters of L-broth (10 g of peptone/liter,
5 g of yeast extract/liter, 5 g of sodium chloride/liter; pH 7.0) at 33°C for 27 hours,
after which cultures were harvested to give 20 g of bacterial cells. From the bacterial
cells obtained, chromosome DNA was extracted according to the Marmur method. To 250
µg of this chromosome DNA, 2 U of
Sau3AI were added, and allowed to react at 37°C for 30 minutes, thereby causing partial
digestion. From the partially digested DNA, 4-9 kbp DNA fragments were obtained by
agarose gel electrophoresis. On the other hand, plasmid pUC_18 was completely digested
with
BamHI, and ligated with T4 DNA ligase to the above-obtained DNA fragments from chromosome,
resulting in a mixed solution of various recombinant plasmids.
Example 2
[0040] Selection of plasmid derived from
Agrobacterium species KNK 712 (FERM BP-1900) and containing gene related to conversion of D-N-carbamoyl-α-amino
acid into the corresponding D-α-amino acid:-
[0041] Using the mixed plasmid solution of Example 1,
Escherichia coli JM 109 was transformed according to the conventional method. This was inoculated
on the medium of Table 1, containing ampicillin as a selection marker.
Table 1
| Polypeptone |
1 g |
| Yeast extract |
0.5 g |
| Sodium chloride |
0.5 g |
| Ampicillin |
100 mg |
| Ager |
15 g |
| Water was added to the volume of 1 liter (pH 7.0). |
[0042] Water was added to the volume of 1 liter (pH 7.0).
[0043] The grown colonies were collected and inoculated into the liquid medium of Table
2, containing D-N-carbamoyl-alanine as a sole nitrogen source, followed by cultivation.
Table 2
| Na2HPO4 |
6 g |
| KH2PO4 |
3 g |
| NaCl |
0.5 g |
| MgSO4 |
0.12 g |
| CaCl2 |
11 mg |
| Glucose |
2 g |
| Ampicillin |
50 mg |
| Thiamin |
1 mg |
| D-N-Carbamoyl-alanine |
1 g |
| Water was added to the volume of 1 liter (pH 7.0). |
[0044] Water was added to the volume of 1 liter (pH 7.0).
[0045] Only the transformed strains expressing the desired gene can be grown on the medium
of Table 2 by utilizing D-N-carbamoyl-alanine as a nitrogen source. The culture solution
in the medium of Table 2 was inoculated into the same medium as used above, and this
operation was repeated for enrichment of the desired transformed strains. From the
enriched culture solution, the transformed strains were purely separated with the
medium of Table 1. These purely separated cells were inoculated into 10 ml of L-broth
(10 g of peptone/liter, 5 g of yeast extract/liter, 5 g of sodium chloride/liter;
pH 7.0) containing 100 mg/liter of ampicillin, and incubated at 37°C for 16 hours,
after which 1 ml of the culture solution was harvested and the supernatant was removed
therefrom, followed by suspension of the bacterial cells in 0.5 ml of substrate solution
(0.5% D-N-carbamoyl-parahydroxyphenylglycine, 0.05% Triton® X-100, 0.1 M phosphate
buffer (hereinafter referred to as KPB); pH 7.0) and allowing to react at 37°C for
3 hours. This reaction mixture was spotted on TLC and developed, followed by staining
with ninhydrin, resulting in a spot of D-parahydroxyphenylglycine which was in agreement
with the standard. From this fact, it was confirmed that the transformed strains purely
separated have a plasmid containing a gene related to the desired enzyme. The plasmid
contained in these strains was named pAHD 101.
[0046] Then, from the above transformed strains, pAHD 101 was prepared in large quantities
according to the alkali denaturation method, followed by mapping, which reveled that
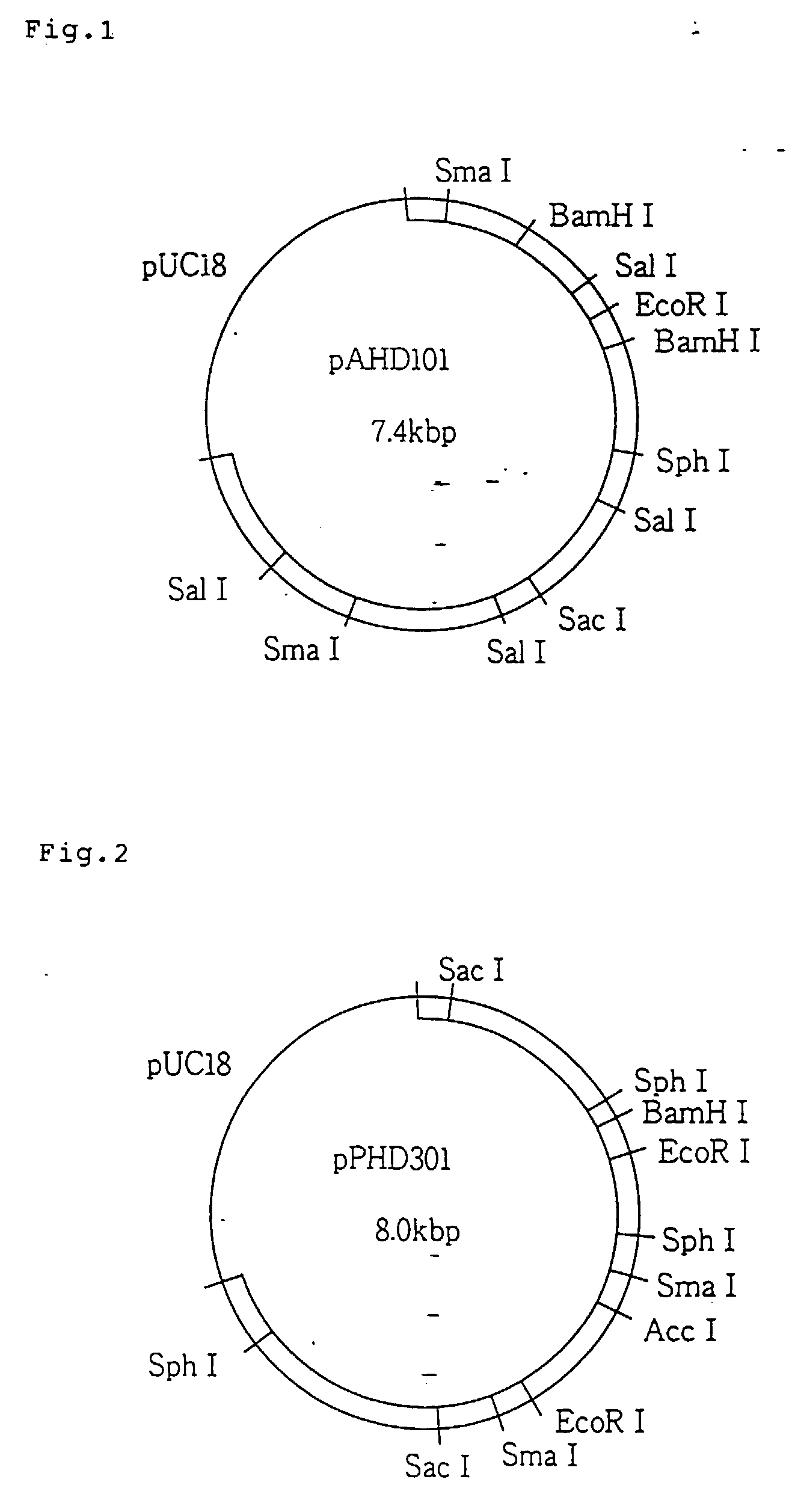
it had the structure shown in Fig. 1.
[0047] Escherichia coli JM 109 transformed with pAHD 101 was named
Escherichia coli JM 109 pAHD 101.
Example 3
[0048] Selection of plasmid derived from
Pseudomonas sp. KNK 003A (FERM BP-3181) and containing gene related to conversion of D-N-carbamoyl-α-amino
acid into the corresponding D-α-amino acid:-
[0049] In the same manner as described in Example 1, a mixed solution of various recombinant
plasmids formed from chromosome DNA fragments of
Pseudomonas sp. KNK 003A (FERM BP-3181) and plasmid pUC18 was obtained. Using this mixed solution,
Escherichia coli JM 109 was transformed according to the conventional method. From these various transformed
strains, particular transformed strains having a plasmid which contains a gene related
to an enzyme capable of converting D-N-carbamoyl-α-amino acids into the corresponding
D-α-amino acids were obtained in the same manner as described in Example 2, except
that the liquid medium of Table 2, containing 100 mg/liter of IPTG, was used in place
of the liquid medium of Table 2. The plasmid contained in these strains was named
pPHD 301.
[0050] Then, from the above transformed strains, pPHD 301 was prepared in large quantities,
followed by mapping, which revealed that it had the structure shown in Fig. 2.
[0051] Escherichia coli JM 109 transformed with pPHD 301 was named
Escherichia coli JM 109 pPHD 301.
Example 4
Subcloning of pAHD 101
[0052] The pAHD 101 obtained in Example 2 was completely digested with
SmaI and
EcoRI, and a 2.7 kbp
SmaI-
EcoRI fragment was obtained by agarose gel electrophoresis. On the other hand, plasmid
pUC 19 was completely digested with
SmaI and
EcoRI, and ligated with T4 DNA ligase to the
SmaI-
EcoRI fragment of pAHD 101, after which
Escherichia coli JM 109 was transformed with this ligated plasmid and the transformed strains were
cultivated on an L-medium (1% peptone, 0.5% yeast extract, 0.5% sodium chloride, 1.5%
agar; pH 7.0) containing isopropyl-1-thio-β-D-galactoside (IPTG), 5-chloro-4-bromo-3-indolyl-β-D-galactose
(Xgal) and ampicillin.
[0053] The plasmid corresponding to pUC 19 containing the
SmaI-
EcoRI fragment of pAHD 101 formed white colonies without exhibiting blue color, and therefore,
these white colonies were selected. To confirm that these selected bacteria have the
desired enzyme activity, they were inoculated into 10 ml of L-broth (1% peptone, 0.5%
yeast extract, 0.5% sodium chloride) containing 100 mg/liter of ampicillin after cultivation
of which 1 ml of the culture solution was harvested and the supernatant was removed
therefrom, and the bacterial cells were suspended in 0.5 ml of substrate solution
(0.5% D-N-carbamoyl-parahydroxyphenylglycine, 0.05% Triton® X-100, 0.1 M phosphate
buffer; pH 7.0), followed by reaction at 37°C for 3 hours. This reaction mixture was
spotted on TLC and developed, followed by staining with ninhydrin, resulting in a
spot of D-parahydroxyphenylglycine which was in agreement with the standard. The plasmid
contained in these transformed strains was named pAD 107.
[0054] Then, the plasmid pAD 107 was prepared and partially hydrolyzed with
SalI, followed by agarose gel electrophoresis to give a 4.5 kbp fragment containing the
pUC 19 portion. This fragment was circularized with T4 DNA ligase, and this plasmid
was used to transform
Escherichia coli JM 109. The trasformed strains were obtained from the L-medium containing ampicillin
and bacterial reaction was conducted in the same manner as described above, thereby
obtaining transformed strains having the ability to convert D-N-carbamoyl-parahydroxyphenylglycine
into D-parahydroxyphenylglycine. The plasmid contained in these transformed strains
was named pAD 108. The pAD 108 was prepared form the above transformed strains, followed
by mapping, which revealed that it had the structure shown in Fig. 3.
[0055] Escherichia coli JM 109 transformed with pAD 108 was named
Escherichia coli JM 109 pAD 108. The accession number given from the Fermentation Research Institute
was FERM BP-3184.
Example 5
Subcloning of pPHD 301
[0056] The pPHD 301 obtained in Example 3 was completely digested with
AccI, and a 5.2 kbp fragment containing the pUC 18 portion was obtained by agarose gel
electrophoresis. This fragment was circularized with T4 DNA ligase, and this plasmid
was used to transform
Escherichia coli JM 109. The transformed strains were obtained from an L-medium containing ampicillin
and bacterial reaction was conducted in the same manner as described in Example 4,
thereby obtaining transformed strains having the ability to convert D-N-carbamoyl-parahydroxyphenylglycine
into D-parahydroxy-phenylglycine. The plasmid contained in these transformed strains
was named pPD 302.
[0057] Then, the plasmid pPD 302 was prepared and completely digested with
SphI and
AccI, after which a 1.8 kbp
SphI-
AccI fragment was obtained by agarose gel electrophoresis. On the other hand, plasmid
pUC 19 was completely digested with
SphI and
AccI, and ligated with T4 DNA ligase to the
SphI-
AccI fragment of pPD 302, after which
Escherichia coli JM 109 was transformed with this ligated plasmid. The trasformed strains were obtained
from an L-medium containing ampicillin, and bacterial reaction was conducted in the
same manner as described in Example 4, thereby obtaining transformed strains having
the ability to convert D-N-carbamoyl-parahydroxyphenylglycine into D-parahydroxyphenylglycine.
The plasmid contained in these transformed strains was named pPD 304. The plasmid
pPD 304 was prepared from the above transformed strains, followed by mapping, which
revealed that it had the structure shown in Fig. 4.
[0058] Escherichia coli JM 109 transformed with pPD 304 was named
Escherichia coli JM 109 pPD 304. The accession number given from the Fermentation Research Institute
was FERM BP-3183.
Example 6
[0059] Conversion of D-N-carbamoyl-α-amino acid into D-α-amino acid with enzyme obtained
from transformed strains:-
[0060] Using the plasmid pAD 108 obtained in Example 4,
Escherichia coli JM 109 was transformed. This strain was cultivated in L-broth containing 100 µg/ml
of ampicillin and 100 µg/ml of IPTG at 37°C for 16 hours. From 100 ml of this culture
solution, bacterial cells were harvested and then suspended in 0.1 M KPB (pH 7.0)
to the volume of 10 ml. This was subjected to ultrasonication while cooling with ice,
followed by centrifugation to give the supernatant as a crude enzyme solution. Using
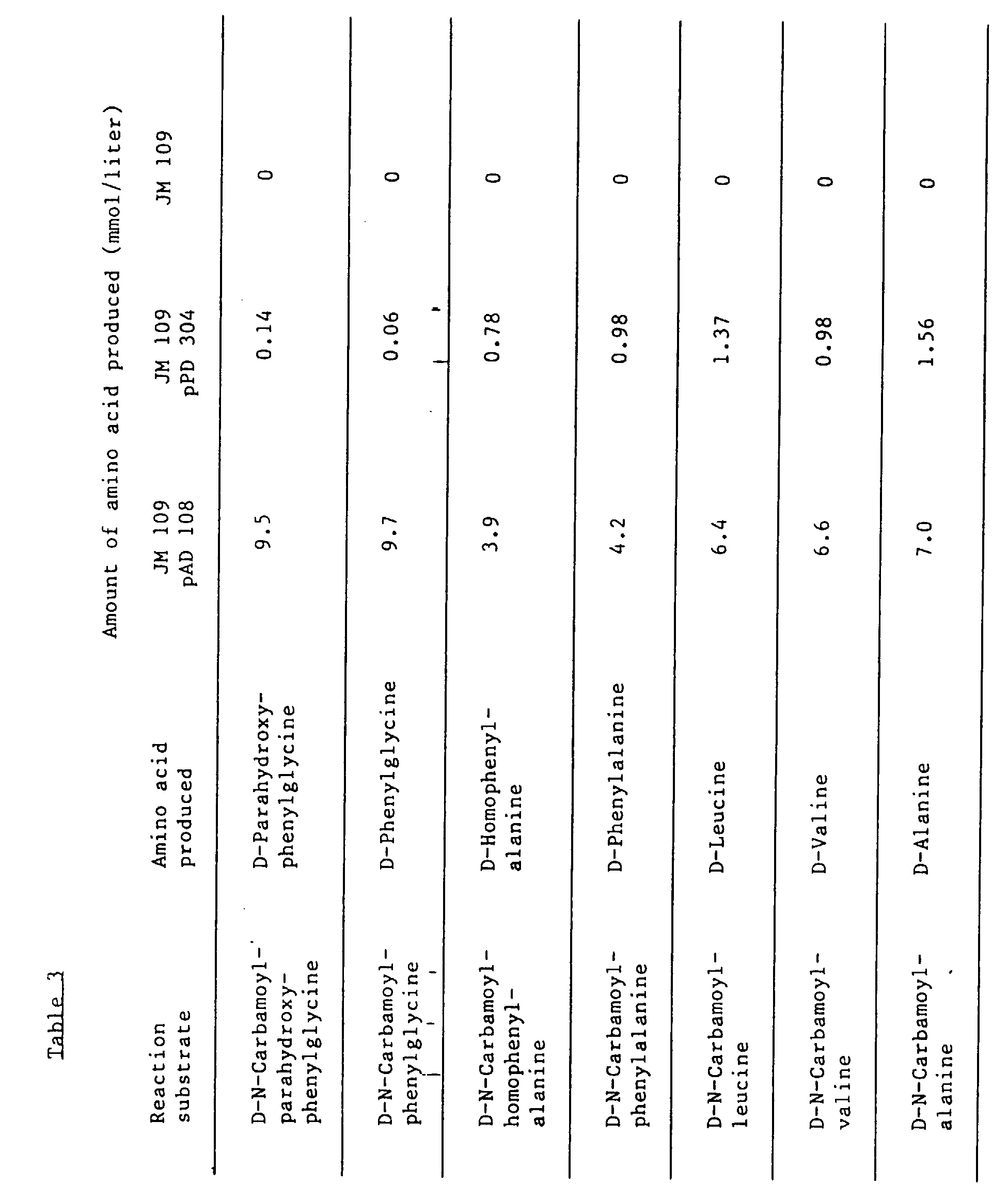
this crude enzyme solution, reaction was conducted with various kinds of D-N-carbamoyl-α-amino
acid shown in Table 3 as a substrate. To 2 ml of 40 mM D-N-carbamoyl-α-amino acid
(Table 3) and 0.2 M KPB (pH 7.0), 100 µl of the above crude enzyme solution were added,
thereby allowing to react at 40°C for 20 minutes, and the amount of D-α-amino acid
produced was determined. The results are shown in Table 3. Moreover, there was a 10-fold
improvement in the specific activity of a D-N-carbamoyl-α-amino acid amidohydrolase
obtained from JM 109 pAD 108 at this time as compared with a crude enzyme solution
from
Agrobacterium species KNK 712 (FERM BP-1900).
Example 7
[0061] Conversion of D-N-carbamoyl-α-amino acid into D-α-amino acid with enzyme obtained
from transformed strains: -
[0062] Using the plasmid pPD 304 obtained in Example 5,
Escherichia coli JM 109 was transformed. From this strain, a crude enzyme solution was obtained in
the same manner as described in Example 6. Using this crude enzyme solution, reaction
was conducted in the same manner as described in Example 6. The results are shown
in Table 3. There was a 40-fold improvement in the specific activity of a D-N-carbamoyl-α-amino
acid amidohydrolase obtained from JM 109 pPD 304 at this time as compared with a crude
enzyme solution from
Pseudomonas sp. KNK 003A (FERM BP-3181).
Example 8
[0063] Conversion of D-N-carbamoyl-parahydroxyphenylglycine into D-parahydroxyphenylglycine
with enzyme obtained from transformed strains:-
[0064] Using the plasmid pAD 108 obtained in Example 4,
Escherichia coli JM 109 was transformed. This strain was cultivated in the same manner as described
in Example 6. From 100 ml of the culture solution, bacterial cells were harvested,
and washed with 0.1 M KPB (pH 7.0), after which they were suspended in 100 ml of 5%
D-N-carbamoyl-parahydroxyphenylglycine, 0.05% Triton® X-100, and 0.1 M KPB (pH 7.0),
followed by stirring at 40°C for 20 hours to cause reaction. The reaction mixture
thus obtained was subjected to centrifugation at 6000 rpm for 10 minutes to remove
the bacterial cells, and pH was decreased to 2.7 by addition of concentrated hydrochloric
acid, followed by adsorption on the cation exchange resin IR-120B (H
+ type) and elution with 5% NH
4OH. Then, the eluates were desalinized with IRC-84 (H
+ type) and decolorized with an AF resin. The decolorized solution was concentrated
to allow crystallization, and the deposited crystals were recrystallized from water
to give 3.8 g of white powder. These crystals exhibited the specific rotation [α]
D20 = -158 (C = 1, 1 N HCl), and gave a single spot on TLC, and the IR spectrum thereof
was in agreement with that of the D-parahydroxyphenylglycine standard.
Example 9
[0065] Preparation of fixed D-N-carbamoyl-α-amino acid amidohydrolase:-
[0066] After 200 ml of the culture solution obtained, in the same manner as described in
Example 6, from the strains of
Escherichia coli JM 109 transformed with pAD 108 was harvested, they were washed with 0.1 M KPB (pH
7.0), and suspended in 20 ml of 0.1 M KPB (pH 7.0), followed by ultrasonication of
the bacterial cells. This suspension of disintegrated bacterial cells was subjected
to centrifugation at 12000 rpm for 20 minutes to give the supernatant as a crude enzyme
solution. To this crude enzyme solution, 2 g of anion exchange resin Duolite A-568
equilibrated with 0.1 M KPB (pH 7.0) were added, and stirred at 4°C for 15 hours to
make the enzyme adsorbed thereon. To this solution, glutaraldehyde was added to have
a final concentration of 0.1%, and stirred for 1 hour, followed by cross-linking treatment,
after which the resin was collected by filtration and washed with 0.1 M KPB, resulting
in 2 g of fixed D-N-carbamoyl-α-amino acid amidohydrolase.
Example 10
[0067] Conversion of D-N-carbamoyl-parahydroxyphenylglycine into D-parahydroxyphenylglycine
with immobilized enzyme:-
[0068] Two grams of immobilized D-N-carbamoyl-α-amino acid amidohydrolase obtained in Example
9 were added to 100 ml of 2% D-N-carbamoyl-parahydroxyphenylglycine and 0.1 M KPB
(pH 7.0), and stirred at 40°C for 20 hours, while maintaining the pH at 7.0 by addition
of 1 N HCl, thereby causing reaction. After the reaction, the mixture was allowed
to stand, and the reaction mixture was collected by suction, afcer which the produced
amino acid was purified in the same manner as described in Example 8 to give 1.5 g
of D-parahydroxyphenylglycine.
Example 11
[0069] Analysis of DNA base sequence for the gene of enzyme derived from
Agrobacterium species KNK 712 (FERM BP-1900) and capable of converting D-N-carbamoyl-α-amino acid into
the corresponding D-α-amino acid:-
[0070] The plasmid pAD 108 containing the gene of a D-N-carbamoyl-α-amino acid amidohydrolase
(hereinafter referred to as amidohydrolase) derived from
Agrobacterium species KNK 712 (FERM BP-1900) was digested with restriction endonucleases
EcoRI and
HindIII (manufactured by Takara Shuzo Co., Ltd.), and a 1.8. kb DNA fragment was separated
for preparation by agarose gel electrophoresis. This fragment was digested with various
restriction endonucleases, and ligated with T4-DNA ligase (manufactured by Takara
Shuzo Co., Ltd.) to M13mp18 or M13mp19, after which
Escherichia coli JM 109 was infected therewith, resulting in the formation of a plaque. This single
plaque was inoculated into 1.5 ml of 2YT medium (16 g/l of bactotrypton (Difco Co.),
10 g/l of yeast extract (Difco Co.), and 5 g/l of NaCl) into which 1% JM 109 had been
inoculated, and subjected to shaking culture at 37°C for 5 hours. After centrifugation,
200 µl of 20% polyethyleneglycol 6000 and 25 M NaCl solution were added to the supernatant,
and allowed to stand at room temperature for 15 minutes, after which phage particles
were recovered as a precipitate by centrifugation. This was dissolved in 100 µl of
TE solution [10 mM Tris HCl (pH 8.0), 1 mM EDTA], and extracted with 50 µl of phenol
(saturated with TE solution), after which 10 µl of 3 M sodium acetate solution and
250 µl of ethanol were added and allowed to stand overnight at -20°C, followed by
centrifugation. After drying, the precipitate was dissolved in 50 µl of TE solution.
Then, 7 µl of this solution was used for reaction, electrophoresis, and autoradiography
with the aid of a DNA sequence kit (manufactured by United States Biochemical Corp.)
using SEQUENASE (registered trade mark) ver. 2, according to its instruction manual.
From the results obtained, the DNA base sequence of amidohydrolase gene for the strain
KNK 712 was determined as shown by SEQ ID No. 1 in the accompanying Sequence Listing.
Example 12
[0071] Analysis of DNA base sequence for the gene of enzyme derived from
Pseudomonas sp. KNK 003A (FERM BP-3181) and capable of converting D-N-carbamoyl-α-amino acid
into the corresponding D-α-amino acid:
[0072] The plasmid pPD 304 containing amidohydrolase gene derived from the strain KNK 003A
was digested with restriction endonucleases
BamHI and
HindIII (manufactured by Takara Shuzo Co., Ltd.), and a 1.8 kb DNA fragment was separated
for preparation by agarose gel electrophoresis. This fragment was digested with various
restriction endonucleases, and ligated with T4-DNA ligase (Takara Shuzo Co., Ltd.)
to M13mpl8 or M13mp19, after which
Escherichia coli JM 109 was infected therewith, resulting in the formation of a plaque. This single
plaque was inoculated into 1.5 ml of 2YT medium (16 g/l bactotrypton (Difco Co.),
10 g/l yeast extract (Difco Co.), and 5 g/l NaCl) into which 1% JM 109 had been inoculated,
and subjected to shaking culture at 37°C for 5 hours. After centrifugation, 200 µl
of 20% polyethyleneglycol 6000 and 25 M NaCl solution were added to the supernatant,
and allowed to stand at room temperature for 15 minutes, after which phage particles
were recovered as a precipitate by centrifugation. This was dissolved in 100 µl of
TE solution [10 mM Tris HCl (pH 8.0), 1 mM EDTA], and extracted with 50 µl of phenol
(saturated with TE solution), after which 10 µl of 3M sodium acetate solution and
250 µl of ethanol were added thereto, and allowed to stand overnight at -20°C, followed
by centrifugation. After drying, the precipitate was dissolved in 50 µl of TE solution.
Then, 7 µl of this solution was used for reaction, electrophoresis, and autoradiography
with the aid of a DNA sequence kit (manufactured by United States Biochemical Corp.)
using SEQUENASE (registered trade mark) ver. 2, according to its instruction mannual.
From the results obtained, the DNA base sequence of amidohydrolase gene derived from
the strain KNK 003A was determined as shown by SEQ ID No. 2 in the accompanying Sequence
Listing.
Example 13
[0073] Purification of D-N-carbamoyl-α-amidohydrolase derived from
Agrobacterium species KNK 712 (FERM BP-1900):-
[0074] Agrobacterium species KNK 712 (FERM BP-1900) was cultivated in the medium of Table 4 at 33°C for 25 hours.
Table 4
| Glycerin |
25 g |
| Sucrose |
5 g |
| KH2PO4 |
5 g |
| Na2HPO4 |
5 g |
| MgSO4.7H2O |
1 g |
| MnCl2.4H2O |
10 mg |
| Yeast extract |
4 g |
| Urea |
2 g |
| D-N-carbamoyl-P-hydroxyphenylglycine |
1 g |
| Water was added to the volume of 1 liter (pH 6.5). |
[0075] Water was added to the volume of 1 liter (pH 6.5).
[0076] Twenty one liters of this culture solution were harvested and the bacterial_cells
were ultrasonicated. After removal of the residue by centrifugation, nucleic acids
were removed by protamine sulfate treatment (0.1 mg/mg of protein). The centrifuged
supernatant was subjected to heat treatment at 50°C for 20 minutes, and after removal
of the precipitate, protein was precipitated by addition of ammonium sulfate, and
a protein fraction having activity and being precipitated with 15% to 35% saturated
ammonium sulfate was recovered. This fraction was dissolved and subjected to HPLC
using DEAE-5pw column (manufactured by Toso Co., Ltd.), followed by elution with a
concentration gradient of NaCl and recovered active fractions. At this stage, in comparison
with a suspension of disintegrated bacterial cells, there was about 20-fold increase
in the specific activity of amidohydrolase. When this fraction was analyzed by SDS-polyacrylamide
gel electrophoresis, this amidohydrolase migrated near to the position corresponding
to the molecular weight of about 35,000.
Example 14
[0077] Determination of amino acid sequence around protein amino terminus of D-N-carbamoyl-α-amino
acid amidohydrolase derived from
Agrobacterium species KNK 712 (FERM BP-1900):-
[0078] A purified amidohydrolase preparation produced by
Agrobacterium species KNK 712 (FERM BP-1900) obtained in Example 13 was charged in a_reverse-phase HPLC
column (AP-303; manufactured by YMC Co.), and eluted with a concentration gradient
of acetonitrile. This fraction containing the amidohydrolase was charged for analysis
in a gas-phase protein sequencer (manufactured by Applied Biosystems Co., Ltd.), and
it was found that the amidohydrolase is a protein having a sequence in the amino terminus
portion, which consists of 1st to 20th amino acids of SEQ ID No. 1 .
Example 15
[0079] Purification of D-N-carbamoyl-α-amino acid amidohydrolase derived from Pseudomonas
sp. KNK 003A (FERM BP-3181):-
[0080] Pseudomonas sp. KNK 003A (FERM BP-3181) was cultivated in the medium of Table 5 at
45°C for 3 days.
Table 5
| Glycerin |
10 g |
| Glucose |
5 g |
| KH2PO4 |
3.5 g |
| Na2HPO4 |
3.5 g |
| MgSO4.7H2O |
0.5 g |
| MnCl2.4H2O |
20 mg |
| FeSO4 |
10 mg |
| CaCO3 |
1 g |
| Meat extract |
2 g |
| Yeast extract |
2 g |
| Polypeptone |
2 g |
| D-N-carbamoylalanine |
1 g |
| Water was added to the volume of 1 liter (pH 7.0). |
[0081] Water was added to the volume of 1 liter (pH 7.0).
[0082] Twenty six liters of this culture solution were harvested, and in the same manner
as described in Example 13, the following operations were conducted: ultrasonication
of the bacterial cells; removal of nucleic acids by protamine sulfate treatment; heat
treatment (65°C, 20 min); fractionation by ammonium sulfate precipitation (separation
of protein fractions having amidohydrolase activity and being precipitated with 50%
to 70% saturated ammonium sulfate); and HPLC using DEAE-5pw column. These active fractions
was allowed to adsorb in a Biogel-HT (Bio-Rad Laboratories Co., Ltd.) column, and
eluted with a concentration gradient of ammonium sulfate, after which the active fractions
were concentrated and subjected to gel filtration using a Sephacryl® S-300 (Pharmacia
LKB Biotechnology Co., Ltd.) column.
[0083] Then, when isoelectric focusing (pH 4 to 6.5) was conducted, the above amidohydrolase
migrated near to the position at pH 5.7, and the gel of this band portion having activity
was cut out, from which protein was extracted. At this stage, in comparison with a
suspension of the disintegrated bacterial cells, there was about 100-fold increase
in the specific activity of amidohydrolase. When this sample was analyzed by SDS-polyacrylamide
gel electrophoresis, amidohydrolase migrated near to the position corresponding to
the molecular weight of about 38,000. Moreover, when this sample was subjected to
gel filtration using a Sephacryl® S-200 column, it was eluted at the position corresponding
to the molecular weight of about 67,000.
Example 16
[0084] Determination of amino acid sequence around protein amino terminus of D-N-carbamoyl-α-amino
acid amidohydrolase derived from
Pseudomonas sp. KNK 003A (FERM BP-3181):-
[0085] A purified amidohydrolase preparation from
Pseudomonas sp. KNK 003A (FERM BP-3181) obtained in Example 15 was charged in a reverse-phase
HPLC column, and eluted with a concentration gradient of acetonitrile. This fraction
containing the amidohydrolase was charged for analysis in a gas-phase protein sequencer,
and it was found that the amidohydrolase is a protein having a sequence in the amino
terminus portion, which consists of 1st to 20th amino acids of SEQ ID No. 2 .
Example 17
[0086] Purification of D-N-carbamoyl-α-amino acid amidohydrolase produced by transformed
Escherichia coli:-
[0087] Escherichia coli JM 109.pAD 108 obtained in Example 4 and
Escherichia coli JM 109.pPD 304 obtained in Example 5 were cultivated by the method as described in
Example 6. The bacterial cells were collected from each culture solution by centrifugation,
and ultrasonicated. After removal of the residue, these crude enzyme solutions were
analyzed by SDS-polyacrylamide electrophoresis. With respect to a suspension of the
cultivated and disintegrated bacterial cells of
Escherichia coli JM 109.pAD 108, the amidohydrolase migrated to the position corresponding to the
molecular weight of about 35,000, and an analysis by densitometer after staining revealed
that the amidohydrolase amounted to about 50% of the whole soluble proteins of the
bacterial cells. Moreover, with respect to a suspension of the cultivated and disintegrated
bacterial cells of
Escherichia coli JM 109.pPD 304, the amidohydrolase migrated to the position corresponding to the
molecular weight of about 38,000, and an analysis by densitometer after staining revealed
that the amidohydrolase amounted to about 15% of the whole soluble proteins of the
bacterial cells.
Example 18
[0088] Determination of protein amino terminal sequence of D-N-carbamoyl-α-amino acid amidohydrolase
produced by transformed
Escherichia coli:-
[0089] A suspension of the cultivated and disintegrated bacterial cells of
Escherichia coli JM 109.pAD 108 obtained in Example 17 was subjected to heat treatment at 50°C for
30 minutes, and after removal of the precipitate by centrifugation, ammonium sulfate
was added to give 30% saturation, thereby causing precipitation of protein. The precipitated
protein was removed by centrifugation and dissolved in deionized water, followed by
desaltization using an NAP-10 column (Pharmacia LKB Biotechnology Co., Ltd.) and charging
in a gas-phase protein sequencer. From the results obtained, it was found that there
were mixed proteins in approximately the same quantities, one having a sequence in
the amino terminus portion, which consists of 1st to 16th amino acids of SEQ ID No.
1, and the other having a sequence in the amino terminus portion, which contains an
additional methionine residue attached to the amino terminus of the former's sequence.
Effect of the invention
[0090] As described hereinabove, the present invention makes it possible to produce D-α-amino
acids from D-N-carbamoyl-α-amino acids with high efficiency by employing gene technology.
Brief Description of the drawings
[0091] Fig. 1 shows the restriction endonuclease map of plasmid pAHD 101 obtained by the
present invention.
[0092] Fig. 2 shows the restriction endonuclease map of plasmid pPHD 301 obtained by the
present invention.
[0093] Fig. 3 shows the restriction endonuclease map of plasmid pAD 108 obtained by the
present invention.
[0094] Fig. 4 shows the restriction endonuclease map of plasmid pPD 304 obtained by the
present invention.
SEQUENCE LISTING
[0095]
SEQ ID NO.: 1
SEQUENCE LENGTH: 1785
SEQUENCE TYPE: nucleic acid
STRANDEDNESS: double
TOPOLOGY: linear
MOLECULE TYPE: genomic DNA
ORIGINAL SOURCE
ORGANISM: Agrobacterium species
STRAIN: KNK 712 (FERM BP-1900)
SEQ ID NO.: 2
[0097]
SEQUENCE LENGTH: 1820
SEQUENCE TYPE: nucleic acid
STRANDEDNESS: double
TOPOLOGY: linear
MOLECULE TYPE: genomic DNA
ORIGINAL SOURCE
ORGANISM: Pseudomonas sp.
STRAIN: KNK 003A (FERM BP-3181)
Claims for the following Contracting State(s): BE, DE, FR, GB, IT, NL
1. A process for the production of D-α-amino acids, by the method in which D-N-carbamoyl-α-amino
acids are converted into the corresponding D-α-amino acids in an aqueous medium with
the aid of an action of an enzyme capable of converting D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids, characterized
in that said enzyme is produced by a transformant which is obtainable by transformation
of host bacterial cells selected from microorganisms belonging to the genus Escherichia,Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium or Brevibacterium with a recombinant DNA comprising a vector DNA and a DNA fragment having a gene encoding
said enzyme, after which the D-α-amino acids produced are collected.
2. A process according to claim 1, wherein D-N-carbamoyl-α-amino acid is a compound of
the general formula:
R-CH(NHCONH2)-COOH
(where R is phenyl, phenyl substituted with hydroxy, alkyl, substituted alkyl, aralkyl,-or
thienyl).
3. A process according to claim 1 or 2, wherein the enzyme produced by the transformant
is present in a culture solution of said transformant, bacterial cells, treated bacterial
cells, extracts from bacterial cells, immobilized bacterial cells, or is used as a
immobilized enzyme.
4. A process according to any one of claims 1 to 3, wherein the DNA fragment containing
said gene is derived from eucaryotes, procaryotes, viruses, bacteriophages, or plasmids.
5. A process according to claim 4, wherein said procaryotes are bacteria.
6. A process according to claim 5, wherein said bacteria are those selected from microorganisms
belonging to the genus Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium, Serratia, or Micrococcus.
7. A process according to claim 6, wherein said bacteria are Pseudomonas sp. KNK 003A (FERM BP-3181) or Pseudomonas sp. KNK 505 (FERM BP-3182).
8. A process according to claim 6, wherein said bacteria are Agrobacterium species KNK 712 (FERM BP-1900).
9. A microorganism including a recombinant comprising a vector DNA and a DNA fragment
having a gene encoding an enzyme capable of converting D-N-carbamoyl-α-amino acids
into the corresponding D-α-amino acids.
10. A microorganism according to claim 9, wherein said microorganism including the recombinant
is one selected from microorganisms belonging to the genus Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium, or Brevibacterium.
11. A microorganism according to claim 10, wherein said microorganism is Escherichia coli JM 109 pAD 108 (FERM BP-3184), or Escherichia coli JM 109 pPD 304 (FERM BP-3183).
12. A process for the production of D-N-carbamoyl-α-amino acid amidohydrolases, characterized
in that a transformant which is obtained by transformation of host bacterial cells
selected from microorganisms belonging to the genus Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium, or Brevibacterium with a recombinant DNA comprising a vector DNA and a DNA fragment having a gene encoding
an enzyme capable of converting D-N-carbamoyl-α-amino acids by removal of their carbamoyl
groups into the corresponding D-α-amino acids is cultivated and said enzyme is recovered.
13. A process according to claim 12, wherein D-N-carbamoyl-α-amino acid is a compound
of the general formula:
R-CH(NHCONH2)-COOH
(where R is phenyl, phenyl substituted hydroxy, alkyl, substituted alkyl, aralkyl,
or thienyl).
14. A recombinant plasmid which is obtained by recombination of plasmid pUC 18 or pUC
19 with a DNA fragment having any one of the restriction endonuclease maps of Figs.
1-4 and containing an gene encoding a D-N-carbamoyl-α-amino acid amidohydrolase derived
from Pseudomonas sp. KNK 003A (FERM BP-3181) or Agrobacterium species KNK 712 (FERM BP-1900).
15. An immobilized enzyme in which an enzyme capable of converting D-N-carbamoyl-α-amino
acids by removal of their carbamoyl groups into the corresponding D-α-amino acids
is fixed on a support for immobilization capable of carrying the enzyme.
16. A gene for a protein having the enzyme activity to convert D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids, said
gene encoding all or a part of the amino acid sequence of 1st to 303rd amino acids
shown in SEQ ID No. 1 in the accompanying Sequence Listing.
17. A DNA fragment in which the base sequence of 167th to 232nd bases of SEQ ID No. 1,
or a base sequence equivalent thereto, is attached to the upstream of the 5' end of
the DNA fragment according to claim 16.
18. A DNA fragment having all or a part of the base sequence of 1st to 1785th bases of
SEQ ID No. 1, or having a base sequence equivalent thereto, and containing a gene
of a protein which has the enzyme activity to convert D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids.
19. A DNA fragment according to claim 18, which comprises a DNA fragment encoding the
amino acid sequence of 1st to 20th amino acids of SEQ ID No. 1.
20. A gene of a protein having the enzyme activity to convert D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids, said
gene encoding all or a part of the amino acid sequence of 1st to 311th amino acids
of SEQ ID No. 2.
21. A DNA fragment having the base sequence of 1st to 1820th bases of SEQ ID No. 2 , or
having a base sequence equivalent thereto, and containing a gene of a protein which
has the enzyme activity to convert D-N-carbamoyl-α-amino acids by removal of their
carbamoyl groups into the corresponding D-α-amino acids.
22. A DNA fragment according to claim 21, which comprises a DNA fragment encoding the
amino acid sequence of 1st to 20th amino acids of SEQ ID No. 2.
23. A protein having all or a part of the amino acid sequence of 1st to 303rd amino acids
of SEQ ID No. 1, and having the enzyme activity to convert D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids.
24. A protein according to claim 23, which has the amino acid sequence of 1st to 20th
amino acids of SEQ ID No. 1.
25. A protein having all or a part of the amino acid sequence of 1st to 311st amino acids
of SEQ ID No. 2, and having the enzyme activity to convert D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids.
26. A protein according to claim 25, which has the amino acid sequence of 1st to 20th
amino acids of SEQ ID No. 2.
27. A microorganism according to claim 9, wherein said vector DNA is a vector which can
autonomously grow in a cell of a microorganism belonging to the genus Escherichia.
28. A process for the production of a D-α-amino acid from a D-N-carbamoyl-α-amino acid
characterized in that an enzyme which removes carbamoyl group of the D-N-carbamoyl-α-amino
acid produced by the microorganism according to claim 9 or 27 to convert it into the
corresponding D-α-amino acid is used.
29. A process for the production of an enzyme for removing carbamoyl group of a D-N-carbamoyl-α-amino
acid to convert it into the corresponding D-α-amino acid characterized in that the
microorganism according to claim 9 or 27 is used.
30. A DNA fragment having all or a part of the base sequence of 230th to 1144th bases
shown in SEQ ID No. 1, or a base sequence equivalent thereto, and containing a gene
encoding a protein which has the enzyme activity to convert a D-N-carbamoyl-α-amino
acid by removal of its carbamoyl group into the corresponding D-α-amino acid.
31. A DNA fragment having all or a part of the base sequence of 233rd to 1141st bases
shown in SEQ ID No. 1, or a base sequence equivalent thereto, and containing a gene
encoding a protein which has the enzyme activity to convert a D-N-carbamoyl-α-amino
acid by removal of its carbamoyl group into the corresponding D-α-amino acid.
32. An expressible recombinant DNA comprising a DNA fragment according to claim 30 or
31 and a vector DNA.
33. A microorganism comprising a recombinant DNA according to claim 32.
34. A process for the production of an enzyme which removes carbamoyl group of a D-N-carbamoyl-α-amino
acid to convert it into the corresponding D-α-amino acid characterized in that a microorganism
according to claim 33 is used.
35. A process for the production of a D-α-amino acid from a D-N-carbamoyl-α-amino acid
characterized in that an enzyme produced by a process according to claim 34 is used.
36. An immobilized enzyme obtained by immobilizing a protein according claim 23 on an
immobilized support.
37. A process for the production of a D-α-amino acid from a D-N-carbamoyl-α-amino acid
characterized in that a protein of claim 23 or an enzyme of claim 36 is used.
38. Use of a protein having decarbamylase activity according to claim 23 for the production
of a D-α-amino acid.
39. Use of an immobilized enzyme according to claim 36 for the production of a D-α-amino
acid.
Claims for the following Contracting State(s): ES
1. A process for the production of D-α-amino acids, by the method in which D-N-carbamoyl-α-amino
acids are converted into the corresponding D-α-amino acids in an aqueous medium with
the aid of an action of an enzyme capable of converting D-N-carbamoyl-α-amino acids
by removal of their carbamoyl groups into the corresponding D-α-amino acids, characterized
in that said enzyme is produced by a transformant which is obtainable by transformation
of host bacterial cells selected from microorganisms belonging to the genus Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium or Brevibacterium with a recombinant DNA comprising a vector DNA and a DNA fragment having a gene encoding
said enzyme, after which the D-α-amino acids produced are collected.
2. A process according to claim 1, wherein D-N-carbamoyl-α-amino acid is a compound of
the general formula:
R-CH(NHCONH2)-COOH
(where R is phenyl, phenyl substituted with hydroxy, alkyl, substituted alkyl, aralkyl,
or thienyl).
3. A process according to claim 1 or 2, wherein the enzyme produced by the transformant
is present in a culture solution of said transformant, bacterial cells, treated bacterial
cells, extracts from bacterial cells, immobilized bacterial cells, or is used as an
immobilized enzyme.
4. A process according to any one of claims 1 to 3, wherein the DNA fragment containing
said gene is derived from eucaryotes, procaryotes, viruses, bacteriophages, or plasmids.
5. A process according to claim 4, wherein said - procaryotes are bacteria.
6. A process according to claim 5, wherein said bacteria are those selected from microorganisms
belonging to the genus Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium,
Serratia, or Micrococcus.
7. A process according to claim 6, wherein said bacteria are Pseudomonas sp. KNK 003A (FERM BP-3181) or Pseudomonas sp. KNK 505 (FERM BP-3182).
8. A process according to claim 6, wherein said bacteria are Agrobacterium species KNK 712 (FERM BP-1900).
9. A process for preparing a microorganism comprising the step of including a recombinant
comprising a vector DNA and a DNA fragment having a gene encoding an enzyme capable
of converting D-N-carbamoyl-α-amino acids into the corresponding D-α-amino acids.
10. A process according to claim 9, wherein said microorganism including the recombinant
is one selected from microorganisms belonging to the genus Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium or Brevibacterium.
11. A process according to claim 10, wherein said microorganism is Escherichia coli JM 109 pAD 108 (FERM BP-3184) or Escherichia coli JM 109 pPD 304 (FERM BP-3183).
12. A process for the production of D-N-carbamoyl-α-amino acid amidohydrolases, characterized
in that a transformant which is obtained by transformation of host bacterial cells
selected from microorganisms belonging to the genus Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium or Brevibacterium with a recombinant DNA comprising a vector DNA and a DNA fragment having a gene encoding
an enzyme capable of converting D-N-carbamoyl-α-amino acids by removal of their carbamoyl
groups into the corresponding D-α-amino acids is cultivated and said enzyme is recovered.
13. A process according to claim 12, wherein D-N-carbamoyl-α-amino acid is a compound
of the general formula:
R-CH(NHCONH2)-COOH
(where R is phenyl, phenyl substituted hydroxy, alkyl, substituted alkyl, aralkyl,
or thienyl).
14. A process of preparing a recombinant plasmid comprising the step of recombining plasmid
pUC 18 or pUC 19 with a DNA fragment having any one of the restriction endonuclease
maps of Figs. 1-4 and containing a gene encoding a D-N-carbamoyl-α-amino acid amidohydrolase
derived from Pseudomonas sp. KNK 003A (FERM BP-3181) or Agrobacterium species KNK 712 (FERM BP 1900).
15. A process for preparing an immobilized enzyme in which an enzyme capable of converting
D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into the corresponding
D-α-amino acids is fixed on a support for immobilization capable of carrying the enzyme.
16. A process for preparing a gene for a protein having the enzyme activity to convert
D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into the corresponding
D-α-amino acids, said gene encoding all or a part of the amino acid sequence of 1st
to 303rd amino acids shown in SEQ ID No. 1, comprising the step of isolating said
gene from Pseudomonas sp. KNK 003A (FERM BP-3181) or Agrobacterium species KNK 712 (FERM BP-1900).
17. A process for preparing a DNA fragment comprising the step of attaching the base sequence
of 167th to 232nd bases of SEQ ID No. 1, or a base sequence equivalent thereto to
the upstream of the 5' end of the DNA fragment according to claim 16.
18. A process for preparing a DNA fragment comprising the step of isolating a DNA sequence,
having all or a part of the base sequence of 1st to 1785th bases of SEQ ID No. 1,
or having a base sequence equivalent thereto, and containing a gene of a protein which
has the enzyme activity to convert D-N-carbamoyl-α-amino acids by removal of their
carbamoyl groups into the corresponding D-α-amino acids.
19. A process according to claim 18, which comprises a DNA fragment encoding the amino
acid sequence of 1st to 20th amino acids of SEQ ID No. 1.
20. A process for preparing a gene of a protein having the enzyme activity to convert
D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into the corresponding
D-α-amino acids, said gene encoding all or a part of the amino acid sequence of 1st
to 311th amino acids of SEQ ID No. 2 comprising the step of isolating said gene from
Pseudomonas sp. KNK 003A (FERM BP-3181) or Agrobacterium species KNK 712 (FERM BP 1900).
21. A process for preparing a DNA fragment, having the base sequence of 1st to 1820th
bases of SEQ ID No. 2, or having a base sequence equivalent thereto, and containing
a gene of a protein which has the enzyme activity to convert D-N-carbamoyl-α-amino
acids by removal of their carbamoyl groups into the corresponding D-α-amino acids,
comprising the step of isolating said DNA sequence.
22. A process according to claim 21, which comprises a DNA fragment encoding the amino
acid sequence of 1st to 20th amino acids of SEQ ID No. 2.
23. A process for preparing a protein having all or a part of the amino acid sequence
of 1st to 303rd amino acids of SEQ ID No. 1, and having the enzyme activity to convert
D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into the corresponding
D-α-amino acids, comprising the step of isolating said protein.
24. A process according to claim 23, wherein the protein has the amino acid sequence of
1st to 20th amino acids of SEQ ID No. 1.
25. A process for preparing a protein having all or a part of the amino acid sequence
of 1st to 311th amino acids of SEQ ID No. 2, and having the enzyme activity to convert
D-N-carbamoyl-α-amino acids by removal of their carbamoyl groups into the corresponding
D-α-amino acids, comprising the step of isolating said protein.
26. A process according to claim 25, which has the amino acid sequence of 1st to 20th
amino acids of SEQ ID No. 2.
27. A process according to claim 9, wherein said vector DNA is a vector which can autonomously
grow in a cell of a microorganism belonging to the genus Escherichia.
28. A process for the production of a D-α-amino acid from a D-N-carbamoyl-α-amino acid
characterized in that an enzyme which removes carbamoyl group of the D-N-carbamoyl-α-amino
acid produced by the microorganism as defined in claim 9 or claim 27 to convert it
into the corresponding D-α-amino acid is used.
29. A process for the production of an enzyme for removing carbamoyl group of a D-N-carbamoyl-α-amino
acid to convert it into the corresponding D-α-amino acid characterized in that the
microorganism according to claim 9 or 27 is used.
30. A process for preparing a DNA fragment having all or a part of the base sequence of
230th to 1144th bases shown in SEQ ID No. 1, or a base sequence equivalent thereto,
and containing a gene encoding a protein which has the enzyme activity to convert
a D-N-carbamoyl-α-amino acid by removal of its carbamoyl group into the corresponding
D-α-amino acid, comprising the step of isolating said DNA fragment.
31. A process for preparing a DNA fragment having all or a part of the base sequence of
233rd to 1141st bases shown in SEQ ID No. 1, or a base sequence equivalent thereto,
and containing a gene encoding a protein which has the enzyme activity to convert
a D-N-carbamoyl-α-amino acid by removal of its carbamoyl group into the corresponding
D-α-amino acid, comprising the step of isolating said DNA fragment.
32. A process for preparing an expressible recombinant DNA comprising the step of inserting
a DNA fragment as defined in claim 30 or claim 31 into a vector DNA.
33. A process for preparing a microorganism comprising the step of including a recombinant
DNA as defined in claim 32.
34. A process for the production of an enzyme which removes carbamoyl group of a D-N-carbamoyl-α-amino
acid to convert it into the corresponding D-α-amino acid characterized in that a microorganism
as defined in claim 33 is used.
35. A process for the production of a D-α-amino acid from a D-N-carbamoyl-α-amino acid
characterized in that an enzyme produced by a process according to claim 34 is used.
36. A process for preparing an immobilized enzyme comprising the step of immobilizing
a protein as defined in claim 23 on an immobilized support.
37. A process for the production of a D-α-amino acid from a D-N-carbamoyl-α-amino acid
characterized in that a protein as defined in claim 23 or an enzyme as defined in
claim 36 is used.
38. Use of a protein having decarbamylase activity as defined in claim 23 for the production
of a D-α-amino acid.
39. Use of a an immobilized enzyme as defined in claim 36 for the production of a D-α-amino
acid.
Patentansprüche für folgende(n) Vertragsstaat(en): BE, DE, FR, GB, IT, NL
1. Verfahren zur Herstellung von D-α-Aminosäuren durch ein Verfahren, worin D-N-Carbamoyl-α-Aminosäuren
in einem wässrigen Medium mit Hilfe der Wirkung eines Enzyms, das D-N-Carbamoyl-α-Aminosäuren
durch Entfernung ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umwandeln
kann, in die entsprechenden D-α-Aminosäuren umgewandelt werden, dadurch gekennzeichnet, daß das Enzym von einem Transformanten hergestellt wird, der erhältlich ist durch
Transformation von Wirtsbakterienzellen, ausgewählt aus den Mikroorganismen, die zur
Gattung Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium oder Brevibacterium gehören, mit einer rekombinanten DNA, umfassend eine Vektor-DNA und ein DNA-Fragment
mit einem dieses Enzym codierenden Gen, woraufhin die erzeugten D-α-Aminosäuren gesammelt
werden.
2. Verfahren nach Anspruch 1, worin D-N-Carbamoyl-α-Aminosäure eine Verbindung der allgemeinen
Formel ist:
R-CH(NHCONH2)-COOH
(worin R Phenyl, Phenyl substituiert mit Hydroxy, Alkyl, substituiertes Alkyl, Aralkyl
oder Thienyl ist).
3. Verfahren nach Anspruch 1 oder 2, worin das von dem Transformanten hergestellte Enzym
in einer Kulturlösung des Transformanten, der Bakterienzellen, der behandelten Bakterienzellen,
der Extrakte aus Bakterienzellen, der immobilisierten Bakterienzellen vorhanden ist
oder als immobilisiertes Enzym verwendet wird.
4. Verfahren nach einem der Ansprüche 1 bis 3, worin das dieses Gen enthaltende DNA-Fragment
von Eukaryonten, Prokaryonten, Viren, Bakteriophagen oder Plasmiden abstammt.
5. Verfahren nach Anspruch 4, worin die Prokaryonten Bakterien sind.
6. Verfahren nach Anspruch 5, worin die Bakterien ausgewählt werden aus Mikroorganismen,
die zur Gattung Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium,
Serratia oder Micrococcus gehören.
7. Verfahren nach Anspruch 6, worin die Bakterien Pseudomonas sp. KNK 003A (FERM BP-3181) oder Pseudomonas sp. KNK 505 (FERM BP-3182) sind.
8. Verfahren nach Anspruch 6, worin die Bakterien die Agrobacterium Art KNK 712 (FERM BP-1900) sind.
9. Mikroorganismus, einschließlich ein rekombinanter, umfassend eine Vektor-DNA und ein
DNA-Fragment mit einem Gen, das ein Enzym codiert, das D-N-Carbamoyl-α-Aminosäuren
in die entsprechenden D-α-Aminosäuren umwandeln kann.
10. Mikroorganismus nach Anspruch 9, worin der Mikroorganismus einschließlich des rekombinanten
ausgewählt ist aus Mikroorganismen, die zur Gattung Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium oder Brevibacterium gehören.
11. Mikroorganismus nach Anspruch 10, worin der Mikroorganismus Escherichia coli JM 109 pAD 108 (FERM BP-3184) oder Escherichia coli JM 109 pPD 304 (FERM BP-3183) ist.
12. Verfahren zur Herstellung von D-N-Carbamoyl-α-Aminosäure Amidohydrolasen, dadurch
gekennzeichnet, daß ein Transformant kultiviert wird, der erhalten wird durch Transformation
von Wirtsbakterienzellen, ausgewählt aus Mikroorganismen, die zur Gattung Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium oder Brevibacterium gehören, mit einer rekombinanten DNA, umfassend eine Vektor-DNA und ein DNA-Fragment
mit einem für ein Enzym codierenden Gen, wobei das Enzym D-N-Carbamoyl-α-Aminosäuren
in die entsprechenden D-α-Aminosäuren umgewandeln kann, und das Enzym gewonnen wird.
13. Verfahren nach Anspruch 12, worin D-N-Carbamoyl-α-Aminosäure eine Verbindung der allgemeinen
Formel ist:
R-CH(NHCONH2)-COOH
(worin R Phenyl, Phenyl substituiert mit Hydroxy, Alkyl, substituiertes Alkyl, Aralkyl
oder Thienyl ist).
14. Rekombinantes Plasmid, das durch Rekombination des Plasmids pUC 18 oder pUC 19 mit
einem DNA-Fragment, das eine der Restriktionsendonukleasen-Karten der Figuren 1 bis
4 hat und ein Gen enthält, das eine von Pseudomonas sp. KNK 003A (FERM BP-3181) oder der Agrobacterium Art KNK 712 (FERM BP-1900) abstammende D-N-Carbamoyl-α-Aminosäure Amidohydrolase
codiert, erhalten wird.
15. Immobilisiertes Enzym, wobei ein Enzym, das D-N-Carbamoyl-α-Aminosäuren durch Entfernen
ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umwandeln kann, auf einem
das Enzym tragenden Träger zur Immobilisation fixiert ist.
16. Gen eines Proteins, das Enzymaktivität hat, um D-N-Carbamoyl-α-Aminosäuren durch Entfernen
ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln, wobei das
Gen die gesamte oder einen Teil der Aminosäuren 1 bis 303 der Aminosäuresequenz, die
in der beiliegenden Sequenzliste (SEQ. ID Nr. 1) gezeigt ist, codiert.
17. DNA-Fragment, worin die Basen 167 bis 232 der Basensequenz der SEQ. ID Nr.1 oder einer
äquivalenten Basensequenz davon stromaufwärts an das 5'-Ende des DNA-Fragments nach
Anspruch 16 gebunden ist.
18. DNA-Fragment, das die gesamten oder einen Teil der Basen 1 bis 1785 der Basensequenz
der SEQ. ID Nr. 1, oder eine äquivalente Basensequenz davon hat, und ein Gen eines
Proteins enthält, das Enzymaktivität hat, um D-N-Carbamoyl-α-Aminosäuren durch Entfernen
ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln.
19. DNA-Fragment nach Anspruch 18, umfassend ein DNA-Fragment, das für die Aminosäuren
1 bis 20 der Aminosäuresequenz der SEQ. ID Nr. 1 codiert.
20. Gen eines Proteins, das Enzymaktivität hat, um D-N-Carbamoyl-α-Aminosäuren durch Entfernen
ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln, wobei das
Gen die gesamte oder einen Teil der Aminosäuren 1 bis 311 der Aminosäuresequenz der
SEQ. ID Nr. 2 codiert.
21. DNA-Fragment, das die Basen 1 bis 1820 der Basensequenz der SEQ. ID Nr. 2, oder eine
äquivalente Basensequenz davon hat, und ein Gen eines Proteins enthält, das Enzymaktivität
hat, um D-N-Carbamoyl-α-Aminosäuren durch Entfernen ihrer Carbamoylgruppen in die
entsprechenden D-α-Aminosäuren umzuwandeln.
22. DNA-Fragment nach Anspruch 21, umfassend ein DNA-Fragment, das für die Aminosäuren
1 bis 20 der Aminosäurensequenz der SEQ. ID Nr. 2 codiert.
23. Protein mit den gesamten oder einem Teil der Aminosäuren 1 bis 303 der Aminosäurensequenz
der SEQ. ID Nr. 1 und mit Enzymaktivität, um D-N-Carbamoyl-α-Aminosäuren durch Entfernen
ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln.
24. Protein nach Anspruch 23, das die Aminosäuren 1 bis 20 der Aminosäuresequenz der SEQ.
ID Nr. 1 hat.
25. Protein mit den gesamten oder einem Teil der Aminosäuren 1 bis 311 der Aminosäuresequenz
der SEQ. ID Nr. 2 und mit Enzymaktivität, um D-N-Carbamoyl-α-Aminosäuren durch Entfernen
ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln.
26. Protein nach Anspruch 25, das die Aminosäuren 1 bis 20 der Aminosäuresequenz der SEQ.
ID Nr. 2 hat.
27. Mikroorganismus nach Anspruch 9, worin die Vektor-DNA ein Vektor ist, der autonom
in einer Zelle eines zur Gattung Escherichia gehörenden Mikroorganismus wächst.
28. Verfahren zur Herstellung von einer D-α-Aminosäure aus einer D-N-Carbamoyl-α-Aminosäure,
dadurch gekennzeichnet, daß ein Enzym verwendet wird, das die Carbamoylgruppe der
D-N-Carbamoyl-α-Aminosäuren entfernt durch einen Mikroorganismus wie in Anspruch 9
oder 27 definiert hergestellt wird, um es in die entsprechende D-α-Aminosäure umzuwandeln.
29. Verfahren zur Herstellung eines Enzyms zum Entfernen der Carbamoylgruppe einer D-N-Carbamoyl-α-Aminosäure,
um es in die entsprechende D-α-Aminosäure umzuwandeln, dadurch gekennzeichnet, daß der Mikroorganismus nach Anspruch 9 oder 27 verwendet wird.
30. DNA-Fragment, das die gesamten oder einen Teil der Basen 230 bis 1144 der Basensequenz,
die in der SEQ. ID Nr. 1 gezeigt ist, oder eine äquivalente Basensequenz davon, hat
und ein Gen enthält, das ein Protein codiert, das Enzymaktivität hat, um eine D-N-Carbamoyl-α-Aminosäure
durch Entfernen ihrer Carbamoylgruppe in die entsprechende D-α-Aminosäure umzuwandeln.
31. DNA-Fragment, das die gesamten oder einen Teil der Basen 233 bis 1141 der Basensequenz,
die in der SEQ. ID Nr. 1 gezeigt ist, oder eine äquivalente Basensequenz davon, hat
und ein Gen enthält, das ein Protein codiert, das Enzymaktivität hat, um eine D-N-Carbamoyl-α-Aminosäure
durch Entfernen ihrer Carbamoylgruppe in die entsprechende D-α-Aminosäure umzuwandeln.
32. Exprimierbare rekombinante DNA, umfassend ein DNA-Fragment nach Anspruch 30 oder 31
und eine Vektor-DNA.
33. Mikroorganismus, umfassend eine rekombinante DNA nach Anspruch 32.
34. Verfahren zur Herstellung eines Enzyms, das eine Carbamoylgruppe von einer D-N-Carbamoyl-α-Aminosäure
entfernt, um sie in die entsprechende D-α-Aminosäure umzuwandeln, dadurch gekennzeichnet,
daß ein Mikroorganismus nach Anspruch 33 verwendet wird.
35. Verfahren zur Herstellung einer D-α-Aminosäure aus einer D-N-Carbamoyl-α-Aminosäure,
dadurch gekennzeichnet, daß ein durch ein Verfahren nach Anspruch 34 hergestelltes Enzym verwendet wird.
36. Immobilisiertes Enzym, das durch Immobilisieren eines Proteins nach Anspruch 23 auf
einem immobilisierten Träger erhalten wird.
37. Verfahren zur Herstellung einer D-α-Aminosäure aus einer D-N-Carbamoyl-α-Aminosäure,
dadurch gekennzeichnet, daß ein Protein nach Anspruch 23 oder ein Enzym nach Anspruch 36 verwendet wird.
38. Verwendung eines Proteins mit Decarbamylase-Aktivität nach Anspruch 23 zur Herstellung
einer D-α-Aminosäure.
39. Verwendung eines immobilisierten Enzyms nach Anspruch 36 zur Herstellung einer D-α-Aminosäure.
Patentansprüche für folgende(n) Vertragsstaat(en): ES
1. Verfahren zur Herstellung von D-α-Aminosäuren durch ein Verfahren, worin D-N-Carbamoyl-α-Aminosäuren
in einem wässrigen Medium mit Hilfe der Wirkung eines Enzyms, das D-N-Carbamoyl-α-Aminosäuren
durch Entfernung ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umwandeln
kann, in die entsprechenden D-α-Aminosäuren umgewandelt werden, dadurch gekennzeichnet, daß das Enzym von einem Transformanten hergestellt wird, der erhältlich ist durch
Transformation von Wirtsbakterienzellen, ausgewählt aus den Mikroorganismen, die zur
Gattung Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium oder Brevibacterium gehören, mit einer rekombinanten DNA, umfassend eine Vektor-DNA und ein DNA-Fragment
mit einem dieses Enzym codierenden Gen, woraufhin die erzeugten D-α-Aminosäuren gesammelt
werden.
2. Verfahren nach Anspruch 1, worin D-N-Carbamoyl-α-Aminosäure eine Verbindung der allgemeinen
Formel ist:
R-CH(NHCONH2)-COOH
(worin R Phenyl, Phenyl substituiert mit Hydroxy, Alkyl, substituiertes Alkyl, Aralkyl
oder Thienyl ist).
3. Verfahren nach Anspruch 1 oder 2, worin das von dem Transformanten hergestellte Enzym
in einer Kulturlösung des Transformanten, der Bakterienzellen, der behandelten Bakterienzellen,
der Extrakte aus Bakterienzellen, der immobilisierten Bakterienzellen vorhanden ist
oder als immobilisiertes Enzym verwendet wird.
4. Verfahren nach einem der Ansprüche 1 bis 3, worin das dieses Gen enthaltende DNA-Fragment
von Eukaryonten, Prokaryonten, Viren, Bakteriophagen oder Plasmiden abstammt.
5. Verfahren nach Anspruch 4, worin die Prokaryonten Bakterien sind.
6. Verfahren nach Anspruch 5, worin die Bakterien ausgewählt werden aus Mikroorganismen,
die zur Gattung Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium,
Serratia oder Micrococcus gehören.
7. Verfahren nach Anspruch 6, worin die Bakterien Pseudomonas sp. KNK 003A (FERM BP-3181) oder Pseudomonas sp. KNK 505 (FERM BP-3182) sind.
8. Verfahren nach Anspruch 6, worin die Bakterien die Agrobacterium Art KNK 712 (FERM BP-1900) sind.
9. Verfahren zum Herstellen eines Mikroorganismus, umfassend den Schritt des Einführens
eines Rekombinanten, umfassend eine Vektor-DNA und ein DNA-Fragment mit einem Gen,
das ein Enzym codiert, das D-N-Carbamoyl-α-Aminosäuren in die entsprechenden D-α-Aminosäuren
umwandeln kann.
10. Verfahren nach Anspruch 9, worin der Mikroorganismus einschließlich des rekombinanten
ausgewählt ist aus Mikroorganismen, die zur Gattung Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium oder Brevibacterium gehören.
11. Verfahren nach Anspruch 10, worin der Mikroorganismus Escherichia coli JM 109 pAD 108 (FERM BP-3184) oder Escherichia coli JM 109 pPD 304 (FERM BP-3183) ist.
12. Verfahren zur Herstellung von D-N-Carbamoyl-α-Aminosäure Amidohydrolasen, dadurch
gekennzeichnet, daß ein Transformant kultiviert wird, der erhalten wird durch Transformation von
Wirtsbakterienzellen, ausgewählt aus Mikroorganismen, die zur Gattung Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium oder Brevibacterium gehören, mit einer rekombinanten DNA, umfassend eine Vektor-DNA und ein DNA-Fragment
mit einem für ein Enzym codierenden Gen, wobei das Enzym D-N-Carbamoyl-α-Aminosäuren
in die entsprechenden D-α-Aminosäuren umgewandeln kann, und das Enzym gewonnen wird.
13. Verfahren nach Anspruch 12, worin D-N-Carbamoyl-α-Aminosäuren eine Verbindung der
allgemeinen Formel ist:
R-CH(NHCONH2)-COOH
(worin R Phenyl, Phenyl substituiert mit Hydroxy, Alkyl, substituiertes Alkyl, Aralkyl
oder Thienyl ist).
14. Verfahren zum Herstellen eines rekombinanten Plasmids, umfassend den Schritt des Rekombinierens
des Plasmids pUC 18 oder pUC 19 mit einem DNA-Fragment, das eine der Restriktionsendonukleasen-Karten
der Figuren 1 bis 4 hat und ein Gen enthält, das eine von Pseudomonas sp. KNK 003A (FERM BP-3181) oder Agrobacterium Arten KNK 712 (FERM BP-1900) abstammende D-N-Carbamoyl-α-Aminosäure Amidohydrolase
codiert.
15. Verfahren zum Herstellen eines immobilisierten Enzyms, worin ein Enzym, das D-N-Carbamoyl-α-Aminosäuren
durch Entfernen ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umwandeln
kann, auf einem Träger, der das Enzym tragen kann, zur Immobilisation fixiert ist.
16. Verfahren zum Herstellen eines Gens von einem Protein mit Enzymaktivität, um D-N-Carbamoyl-α-Aminosäuren
durch Entfernen ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln,
wobei das Gen für die gesamte oder einen Teil der Aminosäuren 1 bis 303 der Aminosäuresequenz,
die in SEQ. ID Nr. 1 gezeigt ist, codiert, umfassend den Schritt des Isolierens des
Gens aus Pseudomonas sp. KNK 003A (FERM BP-3181) oder der Agrobacterium Art KNK 712 (FERM BP-1900).
17. Verfahren zum Herstellen eines DNA-Fragments, umfassend den Schritt des Zusammenfügens
der Basen 167 bis 232 der Basensequenz der SEQ. ID Nr. 1 oder einer äquivalenten Basensequenz
davon stromaufwärts des 5'-Endes des DNA-Fragments nach Anspruch 16.
18. Verfahren zum Herstellen eines DNA-Fragments, umfassend den Schritt des Isolierens
der DNA-Sequenz mit den gesamten oder einem Teil der Basen 1 bis 1785 der Basensequenz
der SEQ. ID Nr. 1, oder mit einer äquivalenten Basensequenz davon, und enthaltend
ein Gen eines Proteins, das Enzymaktivität hat, um D-N-Carbamoyl-α-Aminosäuren durch
Entfernen ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln.
19. Verfahren nach Anspruch 18, umfassend ein DNA-Fragment, das für die Aminosäuren 1
bis 20 der Aminosäuresequenz der SEQ. ID Nr. 1 codiert.
20. Verfahren zum Herstellen eines Gens von einem Protein, das Enzymaktivität hat, um
D-N-Carbamoyl-α-Aminosäuren durch Entfernen ihrer Carbamoylgruppen in die entsprechenden
D-α-Aminosäuren umzuwandeln, wobei das Gen die gesamte oder einen Teil der Aminosäuren
1 bis 311 der Aminosäuresequenz der SEQ. ID Nr. 2 codiert, umfassend den Schritt des
Isolierens des Gens aus Pseudomonas sp. KNK 003A (FERM BP-3181) oder der Agrobacterium Art KNK 712 (FERM BP-1900).
21. Verfahren zum Herstellen eines DNA-Fragments mit den Basen 1 bis 1820 der Basensequenz
der SEQ. ID Nr. 2, oder mit einer äquivalenten Basensequenz davon, und enthaltend
ein Gen eines Proteins, das Enzymaktivität hat, um D-N-Carbamoyl-α-Aminosäuren durch
Entfernen ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln,
umfassend den Schritt des Isolierens der DNA-Sequenz.
22. Verfahren nach Anspruch 21, umfassend ein DNA-Fragment, das für die Aminosäuren 1
bis 20 der Aminosäuresequenz der SEQ. ID Nr. 2 codiert.
23. Verfahren zum Herstellen eines Proteins mit den gesamten oder einem Teil der Aminosäuren
1 bis 303 der Aminosäuresequenz der SEQ. ID Nr. 1 und mit Enzymaktivität, um D-N-Carbamoyl-α-Aminosäuren
durch Entfernen ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln,
umfassend den Schritt des Isolierens des Proteins.
24. Verfahren Anspruch 23, worin das Protein die Aminosäuren 1 bis 20 der Aminosäuresequenz
der SEQ. ID Nr. 1 hat.
25. Verfahren zum Herstellen eines Proteins mit den gesamten oder einem Teil der Aminosäuren
1 bis 311 der Aminosäuresequenz der SEQ. ID Nr. 2 und mit Enzymaktivität, um D-N-Carbamoyl-α-Aminosäuren
durch Entfernen ihrer Carbamoylgruppen in die entsprechenden D-α-Aminosäuren umzuwandeln,
umfassend den Schritt des Isolierens des Proteins.
26. Verfahren nach Anspruch 25, das die Aminosäuren 1 bis 20 der Aminosäuresequenz der
SEQ. ID Nr. 2 hat.
27. Verfahren nach Anspruch 9, worin die Vektor-DNA ein Vektor ist, der autonom in einer
Zelle eines zur Gattung Escherichia gehörenden Mikroorganismus wächst.
28. Verfahren zur Herstellung einer D-α-Aminosäure aus einer D-N-Carbamoyl-α-Aminosäure,
dadurch gekennzeichnet, daß ein Enzym verwendet wird, das die Carbamoylgruppe der
D-N-Carbamoyl-α-Aminosäuren entfernt durch einen Mikroorganismus wie in Anspruch 9
oder 27 definiert hergestellt wird, um es in die entsprechende D-α-Aminosäure umzuwandeln.
29. Verfahren zur Herstellung eines Enzyms zum Entfernen der Carbamoylgruppe von einer
D-N-Carbamoyl-α-Aminosäure, um es in die entsprechende D-α-Aminosäure umzuwandeln,
dadurch gekennzeichnet, daß der Mikroorganismus nach Anspruch 9 oder 27 verwendet wird.
30. Verfahren zum Herstellen eines DNA-Fragments mit den gesamten oder einem Teil der
Basen 230 bis 1144 der Basensequenz, die in SEQ. ID Nr. 1 gezeigt ist, oder einer
äquivalente Basensequenz davon, und enthaltend ein Gen, das ein Protein codiert, das
Enzymaktivität hat, um eine D-N-Carbamoyl-α-Aminosäure durch Entfernen ihrer Carbamoylgruppe
in die entsprechende D-α-Aminosäure umzuwandeln, umfassend den Schritt des Isolierens
des DNA-Fragments.
31. Verfahren zum Herstellen eines DNA-Fragments mit den gesamten oder einem Teil der
Basen 233 bis 1141 der Basensequenz, die in SEQ. ID Nr. 1 gezeigt ist, oder einer
äquivalente Basensequenz davon, und enthaltend ein Gen, das ein Protein codiert, das
Enzymaktivität hat, um eine D-N-Carbamoyl-α-Aminosäure durch Entfernen ihrer Carbamoylgruppe
in die entsprechende D-α-Aminosäure umzuwandeln, umfassend den Schritt des Isolierens
des DNA-Fragments.
32. Verfahren zum Herstellen einer exprimierbaren rekombinanten DNA in eine Vektor-DNA,
umfassend den Schritt des Einführens eines DNA-Fragments, wie in Anspruch 30 oder
31 definiert.
33. Verfahren zum Herstellen eines Mikroorganismus, umfassend den Schritt des Einschließens
einer rekombinanten DNA, wie in Anspruch 32 definiert.
34. Verfahren zur Herstellung eines Enzyms, das eine Carbamoylgruppe von einer D-N-Carbamoyl-α-Aminosäure
entfernt, um sie in entsprechende D-α-Aminosäure umzuwandeln, dadurch gekennzeichnet,
daß ein Mikroorganismus nach Anspruch 33 verwendet wird.
35. Verfahren zur Herstellung einer D-α-Aminosäure aus einer D-N-Carbamoyl-α-Aminosäure,
dadurch gekennzeichnet, daß ein durch ein Verfahren nach Anspruch 34 hergestelltes Enzym verwendet wird.
36. Verfahren zum Herstellen eines immobilisierten Enzyms, umfassend den Schritt des Immobilisierens
eines Proteins auf einem immobilisierten Träger, wie in Anspruch 23 definiert.
37. Verfahren zur Herstellung einer D-α-Aminosäure aus einer D-N-Carbamoyl-α-Aminosäure,
dadurch gekennzeichnet, daß ein Protein, wie in Anspruch 23 definiert, oder ein Enzym,
wie in Anspruch 36 definiert, verwendet wird.
38. Verwendung eines Proteins mit Decarbamylase-Aktivität, wie in Anspruch 23 definiert,
zur Herstellung einer D-α-Aminosäure.
39. Verwendung eines immobilisierten Enzyms, wie in Anspruch 36 definiert, zur Herstellung
einer D-α-Aminosäure.
Revendications pour l'(les) Etat(s) contractant(s) suivant(s): BE, DE, FR, GB, IT,
NL
1. Un procédé pour la production d'acides D-α-aminés, par la méthode dans laquelle des
acides D-N-carbamoyl-α-aminés sont convertis en acides D-α-aminés correspondants dans
un milieu aqueux à l'aide d'une action d'une enzyme capable de convertir des acides
D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides D-α-aminés
correspondants, caractérisé en ce que ladite enzyme est produite par un transformant
qui peut être obtenu par transformation de cellules bactériennes hôtes choisies parmi
des micro-organismes appartenant aux genres Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium ou Brevibacterium avec un ADN recombinant comprenant un ADN vecteur et un fragment d'ADN ayant un gène
codant pour ladite enzyme, après quoi, les acides D-α-aminés produits sont récoltés.
2. Un procédé selon la revendication 1, dans lequel l'acide D-N-carbamoyl-α-aminé est
un composé de formule générale :
R-CH(NHCONH2)-COOH
(dans laquelle R est un phényle, un phényle substitué par un hydroxy, un alkyle, un
alkyle substitué, un aralkyle ou un thiényle).
3. Un procédé selon la revendication 1 ou 2, dans lequel l'enzyme produite par le transformant
est présente dans un milieu de culture liquide dudit transformant, des cellules bactériennes,
des cellules bactériennes traitées, des extraits de cellules bactériennes, des cellules
bactériennes immobilisées, ou est utilisée comme enzyme immobilisée.
4. Un procédé selon l'une quelconque des revendications 1 à 3, dans lequel le fragment
d'ADN contenant ledit gène est obtenu à partir d'eucaryotes, de procaryotes, de virus,
de bactériophages ou de plasmides.
5. Un procédé selon la revendication 4, dans lequel lesdits procaryotes sont des bactéries.
6. Un procédé selon la revendication 5, dans lequel lesdites bactéries sont celles choisies
parmi des micro-organismes appartenant aux genres Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium,
Serratia ou Micrococcus.
7. Un procédé selon la revendication 6, dans lequel lesdites bactéries sont Pseudomonas sp.KNK 003A (FERM BP-3181) ou Pseudomonas sp. KNK 505 (FERM BP-3182).
8. Un procédé selon la revendication 6, dans lequel lesdites bactéries sont Agrobacterium espèce KNK 712 (FERM BP-1900).
9. Un micro-organisme incluant un recombinant comprenant un ADN vecteur et un fragment
d'ADN ayant un gène codant pour une enzyme capable de convertir des acides D-N-carbamoyl-α-aminés
en acides D-α-aminés correspondants.
10. Un micro-organisme selon la revendication 9, dans lequel ledit micro-organisme incluant
le recombinant est un micro-organisme choisi parmi ceux qui appartiennent aux genres
Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium ou Brevibacterium.
11. Un micro-organisme selon la revendication 10, dans lequel ledit micro-organisme est
Escherichia coli JM 109 pAD 108 (FERM BP-3184), ou Escherichia coli JM 109 pPD 304 (FERM BP-3183).
12. Un procédé pour la production d'amidohydrolases d'acides D-N-carbamoyl-α-aminés, caractérisé
en ce qu'un transformant qui est obtenu par transformation de cellules bactériennes
hôtes choisies parmi des micro-organismes appartenant aux genres Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynbacterium, ou Brevibacterium avec un ADN recombinant comprenant un ADN vecteur et un fragment d'ADN ayant un gène
codant pour une enzyme capable de convertir des acides D-N-carbamoyl-α-aminés par
élimination de leurs groupes carbamoyle en acides D-α-aminés correspondants est cultivé
et ladite enzyme est récupérée.
13. Un procédé selon la revendication 12, dans lequel l'acide D-N-carbamoyl-α-aminé est
un composé de formule générale :
R-CH(NHCONH2)-COOH
(dans laquelle R est un phényle, un phényle substitué par un hydroxy, un alkyle, un
alkyle substitué, un aralkyle ou un thiényle).
14. Un plasmide recombinant qui est obtenu par recombinaison du plasmide pUC 18 ou pUC
19 avec un fragment d'ADN ayant l'une quelconque des cartes de restriction endonucléasique
des Figures 1-4 et contenant un gène codant pour une aminohydrolase d'acides D-N-carbamoyl-α-aminés
provenant de Pseudomonas sp. KNK 003A (FERM BP-3181) ou Agrobacterium espèce KNK 712 (FERM BP-1900).
15. Une enzyme immobilisée dans laquelle une enzyme capable de convertir des acides D-N-carbamoyl-α-aminés
par élimination de leurs groupes carbamoyle en acides D-α-aminés correspondants est
fixée sur un support pour immobilisation capable de porter l'enzyme.
16. Un gène pour une protéine ayant l'activité enzymatique consistant à convertir des
acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants, ledit gène codant pour toute ou une partie de la séquence
d'acides aminés du 1er au 303ème acide aminé indiquée dans SEQ ID N° 1 dans le listing des séquences ci-joint.
17. Un fragment d'ADN dans lequel la séquence de bases de la 167ème à la 232ème base dans SEQ ID N° 1, ou une séquence de bases équivalente à celle-ci, est attachée
en amont de l'extrémité 5' du fragment d'ADN selon la revendication 16.
18. Un fragment d'ADN ayant toute ou une partie de la séquence de bases de la 1ère à la 1785ème base dans SEQ ID N° 1, ou ayant une séquence de bases équivalente à celle-ci, et
contenant un gène d'une protéine qui a l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants.
19. Un fragment d'ADN selon la revendication 18, qui comprend un fragment d'ADN codant
pour la séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 1.
20. Un gène d'une protéine ayant l'activité enzymatique consistant à convertir des acides
D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides D-α-aminés
correspondants, ledit gène codant pour toute ou une partie de la séquence d'acides
aminés du 1er au 311ème acide aminé dans SEQ ID N° 2.
21. Un fragment d'ADN ayant la séquence de bases de la 1ère à la 1820ème base dans SEQ ID N° 2, ou ayant une séquence de bases équivalente à celle-ci, et
contenant un gène d'une protéine qui a l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants.
22. Un fragment d'ADN selon la revendication 21, qui comprend un fragment d'ADN codant
pour la séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 2.
23. Une protéine ayant toute ou une partie de la séquence d'acides aminés du 1er au 303ème acide aminé dans SEQ ID N° 1, et ayant l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants.
24. Une protéine selon la revendication 23, qui a la séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 1.
25. Une protéine ayant toute ou une partie de la séquence d'acides aminés du 1er au 311ème acide aminé dans SEQ ID N° 2, et ayant l'activité enzymatique consisant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants.
26. Une protéine selon la revendication 25, qui a la séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 2.
27. Un micro-organisme selon la revendication 9, dans lequel ledit ADN vecteur est un
vecteur qui peut se multiplier de manière autonome dans une cellule d'un micro-organisme
appartenant au genre Escherichia.
28. Un procédé pour la production d'un acide D-α-aminé à partir d'un acide D-N-carbamoyl-α-aminé
caractérisé en ce qu'une enzyme qui élimine le groupe carbamoyle de l'acide D-N-carbamoyl-α-aminé
produite par le micro-organisme selon la revendication 9 ou 27 pour le convertir en
acide D-α-aminé correspondant est utilisée.
29. Un procédé pour la production d'une enzyme pour l'élimination du groupe carbamoyle
d'un acide D-N-carbamoyl-α-aminé pour le convertir en acide D-α-aminé correspondant
caractérisé en ce que le micro-organisme selon la revendication 9 ou 27 est utilisé.
30. Un fragment d'ADN ayant toute ou une partie de la séquence de bases de la 230ème à la 1144ème base indiquée dans SEQ ID N° 1, ou une séquence de bases équivalente à celle-ci,
et contenant un gène codant pour une protéine qui a l'activité enzymatique consistant
à convertir un acide D-N-carbamoyl-α-aminé par élimination de son groupe carbamoyle
en acide D-α-aminé correspondant.
31. Un fragment d'ADN ayant toute ou une partie de la séquence de bases de la 233ème à la 1141ème base présentée dans SEQ ID N° 1, ou une séquence de bases équivalente à celle-ci,
et contenant un gène codant pour une protéine qui a l'activité enzymatique consistant
à convertir un acide D-N-carbamoyl-α-aminé par élimination de son groupe carbamoyle
en acide D-α-aminé correspondant.
32. Un ADN recombinant capable de s'exprimer comprenant un fragment d'ADN selon la revendication
30 ou 31 et un ADN vecteur.
33. Un micro-organisme comprenant un ADN recombinant selon la revendication 32.
34. Un procédé pour la production d'une enzyme qui élimine le groupe carbamoyle d'un acide
D-N-carbamoyl-α-aminé pour le convertir en acide D-α-aminé correspondant caractérisé
en ce qu'un micro-organisme selon la revendication 33 est utilisé.
35. Un procédé pour la production d'un acide D-α-aminé à partir d'un acide D-N-carbamoyl-α-aminé
caractérisé en ce qu'une enzyme produite par un procédé selon la revendication 34
est utilisée.
36. Une enzyme immobilisée obtenue par immobilisation d'une protéine selon la revendication
23 sur un support immobilisé.
37. Un procédé pour la production d'un acide D-α-aminé à partir d'un acide D-N-carbamoyl-α-aminé
caractérisé en ce qu'une protéine de la revendication 23 ou une enzyme de la revendication
36 est utilisée.
38. Utilisation d'une protéine ayant une activité décarbamylasique selon la revendication
23 pour la production d'un acide D-α-aminé.
39. Utilisation d'une enzyme immobilisée selon la revendication 36 pour la production
d'un acide D-α-aminé.
Revendications pour l'(les) Etat(s) contractant(s) suivant(s): ES
1. Un procédé pour la production d'acides D-α-aminés, par la méthode dans laquelle des
acides D-N-carbamoyl-α-aminés sont convertis en acides D-α-aminés correspondants dans
un milieu aqueux à l'aide d'une action d'une enzyme capable de convertir des acides
D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides D-α-aminés
correspondants, caractérisé en ce que ladite enzyme est produite par un transformant
qui peut être obtenu par transformation de cellules bactériennes hôtes choisies parmi
des micro-organismes appartenant aux genres Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium ou Brevibacterium avec un ADN recombinant comprenant un ADN vecteur et un fragment d'ADN ayant un gène
codant pour ladite enzyme, après quoi, les acides D-α-aminés produits sont récoltés.
2. Un procédé selon la revendication 1, dans lequel l'acide D-N-carbamoyl-α-aminé est
un composé de formule générale :
R-CH(NHCONH2)-COOH
(dans laquelle R est un phényle, un phényle substitué par un hydroxy, un alkyle, un
alkyle substitué, un aralkyle, ou un thiényle).
3. Un procédé selon la revendication 1 ou 2, dans lequel l'enzyme produite par le transformant
est présente dans un milieu de culture liquide dudit transformant, des cellules bactériennes,
des cellules bactériennes traitées, des extraits de cellules bactériennes, des cellules
bactériennes immobilisées, ou est utilisé comme enzyme immobilisée.
4. Un procédé selon l'une quelconque des revendications 1 à 3, dans lequel le fragment
d'ADN contenant ledit gène est obtenu à partir d'eucaryotes, de procaryotes, de virus,
de bactériophages, ou de plasmides.
5. Un procédé selon la revendication 4, dans lequel lesdits procaryotes sont des bactéries.
6. Un procédé selon la revendication 5, dans lequel lesdites bactéries sont celles choisies
parmi des micro-organismes appartenant aux genres Pseudomonas, Agrobacterium, Aerobacter, Aeromonas, Brevibacterium, Bacillus, Flavobacterium, Serratia, ou Micrococcus.
7. Un procédé selon la revendication 6, dans lequel lesdites bactéries sont Pseudomonas sp. KNK 003A (FERM BP-3181) ou Pseudomonas sp. KNK 505 (FERM BP-3182).
8. Un procédé selon la revendication 6, dans lequel lesdites bactéries sont Agrobacterium espèce KNK 712 (FERM BP-1900).
9. Un procédé pour préparer un micro-organisme comprenant l'étape consistant à inclure
un recombinant comprenant un ADN vecteur et un fragment d'ADN ayant un gène codant
pour une enzyme capable de convertir des acides D-N-carbamoyl-α-aminés en acides D-α-aminés
correspondants.
10. Un procédé selon la revendication 9, dans lequel ledit micro-organisme incluant le
recombinant est un micro-organisme choisi parmi ceux qui appartiennent aux genres
Escherichia, Pseudomonas, Flavobacterium, Bacillus, Serratia, Corynebacterium ou Brevibacterium.
11. Un procédé selon la revendication 10, dans lequel ledit micro-organisme est Escherichia coli JM 109 pAD 108 (FERM BP-3184) ou Escherichia coli JM 109 pPD 304 (FERM BP-3183).
12. Un procédé pour la production d'amidohydrolases d'acide D-N-carbamoyl-α-aminés, caractérisé
en ce qu'un transformant qui est obtenu par transformation de cellules bactériennes
hôtes choisies parmi des micro-organismes appartenant aux genres Escherichia, Pseudomonas, Flavobacterium, Bacillus, sSerratia, Corynebacterium ou Brevibacterium avec un ADN recombinant comprenant un ADN vecteur et un fragment d'ADN ayant un gène
codant pour une enzyme capable de convertir des acides D-N-carbamoyl-α-aminés par
élimination de leurs groupes carbamoyle en acides D-α-aminés correspondants est cultivé
et ladite enzyme est récupérée.
13. Un procédé selon la revendication 12, dans lequel l'acide D-N-carbamoyl-α-aminé est
un composé de formule générale :
R-CH(NHCONH2)-COOH
(dans laquelle R est un phényle, un phényle substitué par un hydroxy, un alkyle substitué,
un alkyle, aralkyle ou unthiényle).
14. Un procédé pour préparer un plasmide recombinant comprenant l'étape consistant à recombiner
un plasmide pUC 18 ou pUC 19 avec un fragment d'ADN ayant l'une quelconque des cartes
de restriction endonucléasique des Figures 1-4 et contenant un gène codant pour une
amidohydrolase d'acides D-N-carbamoyl-α-aminés provenant de Pseudomonas sp. KNK 003A (FERM BP-3181) ou Agrobacterium espèce KNK 712 (FERM BP-1900).
15. Un procédé pour préparer une enzyme immobilisée dans lequel une enzyme capable de
convertir des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle
en acides D-α-aminés correspondants est fixée sur un support pour immobilisation capable
de porter l'enzyme.
16. Un procédé pour préparer un gène pour une protéine ayant l'activité enzymatique consistant
à convertir des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle
en acides D-α-aminés correspondants, ledit gène codant pour toute ou une partie de
la séquence d'acides aminés du 1er au 303ème acide aminé présentée dans SEQ ID N° 1, comprenant l'étape consistant à isoler ledit
gène à partir de Pseudomonas sp. KNK 003A (FERM BP-3181) ou Agrobacterium espèce KNK 712 (FERM BP-1900).
17. Un procédé pour préparer un fragment d'ADN comprenant l'étape consistant à attacher
la séquence de bases de la 167ème à la 232ème base dans SEQ ID N° 1, ou une séquence de bases équivalente à celle-ci, en amont
de l'extrémité 5' du fragment d'ADN selon la revendication 16.
18. Un procédé pour préparer un fragment d'ADN comprenant l'étape consistant à isoler
une séquence d'ADN, ayant toute ou une partie de la séquence de bases de la 1ère à la 1785ème base dans SEQ ID N° 1, ou ayant une séquence de bases équivalente à celle-ci, et
contenant un gène d'une protéine qui a l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants.
19. Un procédé selon la revendication 18, qui comprend un fragment d'ADN codant pour la
séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 1.
20. Un procédé pour préparer un gène d'une protéine ayant l'activité enzymatique consistant
à convertir des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle
en acides D-α-aminés correspondants, ledit gène codant pour toute ou une partie de
la séquence d'acides aminés du 1er au 311ème acide aminé dans SEQ ID N° 2 comprenant l'étape consistant à isoler ledit gène à
partir de Pseudomonas sp. KNK 003A (FERM BP-3181) ou Agrobacterium espèce KNK 712 (FERM BP-1900).
21. Un procédé pour préparer un fragment d'ADN, ayant la séquence de bases de la 1ère à la 1820ème base dans SEQ ID N° 2, ou ayant une séquence de bases équivalente à celle-ci, et
contenant un gène d'une protéine qui a l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants, comprenant l'étape consistant à isoler ladite séquence
d'ADN.
22. Un procédé selon la revendication 21, qui comprend un fragment d'ADN codant pour la
séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 2.
23. Un procédé pour préparer une protéine ayant toute ou une partie de la séquence d'acides
aminés du 1er au 303ème acide aminé dans SEQ ID N° 1, et ayant l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants, comprenant l'étape consistant à isoler ladite protéine.
24. Un procédé selon la revendication 23, dans lequel la protéine a la séquence d'acides
aminés du 1er au 20ème acide aminé dans SEQ ID N° 1.
25. Un procédé pour préparer une protéine ayant toute ou une partie de la séquence d'acides
aminés du 1er au 303ème acide aminé dans SEQ ID N° 2, et ayant l'activité enzymatique consistant à convertir
des acides D-N-carbamoyl-α-aminés par élimination de leurs groupes carbamoyle en acides
D-α-aminés correspondants, comprenant l'étape consistant à isoler ladite protéine.
26. Un procédé selon la revendication 25, qui a la séquence d'acides aminés du 1er au 20ème acide aminé dans SEQ ID N° 2.
27. Un procédé selon la revendication 9, dans lequel ledit ADN vecteur est un vecteur
qui peut se multiplier de manière autonome dans une cellule d'un micro-organisme appartenant
au genre Escherichia.
28. Un procédé pour la production d'un acide D-α-aminé à partir d'un acide D-N-carbamoyl-α-aminé
caractérisé en ce qu'une enzyme qui élimine le groupe carbamoyle de l'acide D-N-carbamoyl-α-aminé
produite par le micro-organisme tel que défini à la revendication 9 ou à la revendication
27 pour le convertir en acide D-α-aminé correspondant est utilisée.
29. Un procédé pour la production d'une enzyme pour l'élimination du groupe carbamoyle
d'un acide D-N-carbamoyl-α-aminé pour le convertir en acide D-α-aminé correspondant
caractérisé en ce que le micro-organisme selon la revendication 9 ou 27 est utilisé.
30. Un procédé pour préparer un fragment d'ADN ayant toute ou une partie de la séquence
de bases de la 230ème à la 1144ème base présentée dans SEQ ID N° 1, ou une séquence de bases équivalente à celle-ci,
et contenant un gène codant pour une protéine qui a l'activité enzymatique consistant
à convertir un acide D-N-carbamoyl-α-aminé par élimination de son groupe carbamoyle
en acide D-α-aminé correspondant, comprenant l'étape consistant à isoler ledit fragment
d'ADN.
31. Un procédé pour préparer un fragment d'ADN ayant toute ou une partie de la séquence
de bases de la 233ème à la 1141ème base présentée dans SEQ ID N° 1, ou une séquence de bases équivalente à celle-ci,
et contenant un gène codant pour une protéine qui a l'activité enzymatique consistant
à convertir un acide D-N-carbamoyl-α-aminé par élimination de son groupe carbamoyle
en acide D-α-aminé correspondant, comprenant l'étape consistant à isoler ledit fragment
d'ADN.
32. Un procédé pour préparer un ADN recombinant capable de s'exprimer comprenant l'étape
consistant à insérer un fragment d'ADN tel que défini à la revendication 30 ou à la
revendication 31 dans un ADN vecteur.
33. Un procédé pour préparer un micro-organisme comprenant l'étape consistant à inclure
un ADN recombinant tel que défini à la revendication 32.
34. Un procédé pour la production d'une enzyme qui élimine le groupe carbamoyle d'un acide
D-N-carbamoyl-α-aminé pour le convertir en acide D-α-aminé correspondant caractérisé
en ce qu'un micro-organisme tel que défini à la revendication 33 est utilisée.
35. Un procédé pour la production d'un acide D-α-aminé à partir d'un acide D-N-carbamoyl-α-aminé
caractérisé en ce qu'une enzyme produite par un procédé selon la revendication 34
est utilisé.
36. Un procédé pour préparer une enzyme immobilisée comprenant l'étape consistant à immobiliser
une protéine telle que définie à la revendication 23 sur un support immobilisé.
37. Un procédé pour la production d'un acide D-α-aminé à partir d'un acide D-N-carbamoyl-α-aminé
caractérisé en ce qu'une protéine telle que définie à la revendication 23 ou une enzyme
telle que définie à la revendication 36 est utilisée.
38. Utilisation d'une protéine ayant une activité décarbamylasique telle que définie à
la revendication 23 pour la production d'un acide D-α-aminé.
39. Utilisation d'une enzyme immobilisée telle que définie à la revendication 36 pour
la production d'un acide D-α-aminé.